Abstract
Progressive neuronal loss and brain atrophy are principal determinants of cognitive decline in Alzheimer’s disease (AD), yet most mouse models fail to recapitulate these features. Here we identify cyclin-dependent kinase 3 (CDK3) as a key driver of neurodegeneration in AD. CDK3 is elevated in human AD brains and correlates with disease severity. As laboratory mice carry a nonfunctional Cdk3 mutation, we generated two models with restored CDK3 activity and then crossed to AD backgrounds. Both models exhibited substantial neuronal loss, hippocampal atrophy, and cognitive deficits. Single-nucleus RNA sequencing revealed neurons simultaneously activating cell cycle and cell death pathways, indicating cell cycle reentry-induced death. Proteomics implicated synaptic proteins as CDK3 substrates. Finally, we developed BMX330, an inhibitor of CDK3, which mitigated neuronal death and rescued cognitive decline in CDK3-restored AD mice. These findings position CDK3 as a therapeutic target and provide pathologically relevant AD models.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The snRNA-seq data are available at the Genome Sequence Archive under the project accession number PRJCA022223 and are accessible at https://ngdc.cncb.ac.cn/gsa/browse/CRA014079. The proteomic data have been successfully submitted to the ProteomeXchange via the PRIDE database, with the following project accession numbers: PXD048032 for the CDK3-interactome (IP), PXD046376 for the proteome and phosphoproteome, and PXD063971 for the APEX2-based CDK3-Interactome in cultured neurons. All other data in this study supporting the findings are available within the article and Supplementary Information files. Materials and reagents used in this study are listed in the Nature Portfolio Reporting Summary. Reagents generated in our laboratory in this study or previous studies are available upon request. Further information including resources and reagents are available from the corresponding author upon reasonable request.
References
Long, J. M. & Holtzman, D. M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
van Dyck, C. H. et al. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med 388, 9–21 (2023).
Largent, E. A., Peterson, A. & Lynch, H. F. FDA drug approval and the ethics of desperation. JAMA Intern. Med. 181, 1555–1556 (2021).
Caselli, R. J., Beach, T. G., Knopman, D. S. & Graff-Radford, N. R. Alzheimer disease: scientific breakthroughs and translational challenges. Mayo Clin. Proc. 92, 978–994 (2017).
Pini, L. et al. Brain atrophy in Alzheimer’s disease and aging. Ageing Res. Rev. 30, 25–48 (2016).
Zahs, K. R. & Ashe, K. H. ‘Too much good news’ - are Alzheimer mouse models trying to tell us how to prevent, not cure, Alzheimer’s disease?. Trends Neurosci. 33, 381–389 (2010).
Gotz, J., Bodea, L. G. & Goedert, M. Rodent models for Alzheimer disease. Nat. Rev. Neurosci. 19, 583–598 (2018).
Balusu, S. et al. MEG3 activates necroptosis in human neuron xenografts modeling Alzheimer’s disease. Science 381, 1176–1182 (2023).
King, A. The search for better animal models of Alzheimer’s disease. Nature 559, S13–S15 (2018).
Jawhar, S., Trawicka, A., Jenneckens, C., Bayer, T. A. & Wirths, O. Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Abeta aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 33, 196 e129–140 (2012).
Herrup, K. & Yang, Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 8, 368–378 (2007).
Nagy, Z., Esiri, M. M., Cato, A. M. & Smith, A. D. Cell cycle markers in the hippocampus in Alzheimer’s disease. Acta Neuropathol. 94, 6–15 (1997).
Yang, Y., Geldmacher, D. S. & Herrup, K. DNA replication precedes neuronal cell death in Alzheimer’s disease. J. Neurosci. 21, 2661–2668 (2001).
Hochegger, H., Takeda, S. & Hunt, T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all?. Nat. Rev. Mol. Cell Biol. 9, 910–U926 (2008).
Welch, P. J. & Wang, J. Y. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell 75, 779–790 (1993).
Ren, S. J. & Rollins, B. J. Cyclin C/Cdk3 promotes Rb-dependent G0 exit. Cell 117, 239–251 (2004).
Braun, K. et al. Investigation of the cell cycle regulation of cdk3-associated kinase activity and the role of cdk3 in proliferation and transformation. Oncogene 17, 2259–2269 (1998).
Hofmann, F. & Livingston, D. M. Differential effects of cdk2 and cdk3 on the control of pRb and E2F function during G1 exit. Genes Dev. 10, 851–861 (1996).
Ye, X., Zhu, C. & Harper, J. W. A premature-termination mutation in the Mus musculus cyclin-dependent kinase 3 gene. Proc. Natl Acad. Sci. USA 98, 1682–1686 (2001).
Dragatsis, I. & Zeitlin, S. CaMKIIalpha-Cre transgene expression and recombination patterns in the mouse brain. Genesis 26, 133–135 (2000).
Radde, R. et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7, 940–946 (2006).
Yoshiyama, Y. et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 (2007).
Lu, T. et al. Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 (2004).
Zhang, B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med 26, 131–142 (2020).
Bai, B. et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron 106, 700 (2020).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Tang, X. Y. et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Invest. 131, 1–16 (2021).
Gui, W. et al. Structural basis of CDK3 activation by cyclin E1 and inhibition by dinaciclib. Biochem. Biophys. Res. Commun. 662, 126–134 (2023).
Ren, S. & Rollins, B. J. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell 117, 239–251 (2004).
Fleisher, A. S. et al. Volumetric MRI vs clinical predictors of Alzheimer disease in mild cognitive impairment. Neurology 70, 191–199 (2008).
Teipel, S. J. et al. Relevance of magnetic resonance imaging for early detection and diagnosis of Alzheimer disease. Med. Clin. North Am. 97, 399–424 (2013).
Iqbal, K., Liu, F., Gong, C. X. & Grundke-Iqbal, I. Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 7, 656–664 (2010).
Bloom, G. S. Amyloid-beta and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 71, 505–508 (2014).
Goedert, M., Spillantini, M. G., Cairns, N. J. & Crowther, R. A. Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168 (1992).
Spillantini, M. G., Crowther, R. A., Kamphorst, W., Heutink, P. & van Swieten, J. C. Tau pathology in two Dutch families with mutations in the microtubule-binding region of tau. Am. J. Pathol. 153, 1359–1363 (1998).
Fujita, K. et al. Targeting Tyro3 ameliorates a model of PGRN-mutant FTLD-TDP via tau-mediated synaptic pathology. Nat. Commun. 9, 433 (2018).
Saito, T. et al. Cdk5 increases MARK4 activity and augments pathological tau accumulation and toxicity through tau phosphorylation at Ser262. Hum. Mol. Genet. 28, 3062–3071 (2019).
Noble, W. et al. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron 38, 555–565 (2003).
Zhang, W., Chen, H., Qu, X., Chang, C. P. & Shou, W. Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC). Am. J. Med. Genet. C 163C, 144–156 (2013).
Fang, X. et al. Loss of cardiac ferritin H facilitates cardiomyopathy via Slc7a11-mediated ferroptosis. Circ. Res. 127, 486–501 (2020).
Kovács, G. G. et al. Genetic prion disease: the EUROCJD experience. Hum. Genet. 118, 166–174 (2005).
Singh, S., Vanden Broeck, A., Miller, L., Chaker-Margot, M. & Klinge, S. Nucleolar maturation of the human small subunit processome. Science 373, 1–13 (2021).
Okuno, Y., McNairn, A. J., den Elzen, N., Pines, J. & Gilbert, D. M. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20, 4263–4277 (2001).
Tang, H. et al. MDM2 is implicated in high-glucose-induced podocyte mitotic catastrophe via Notch1 signalling. J. Cell. Mol. Med. 21, 3435–3444 (2017).
Taipale, M. et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat. Biotechnol. 31, 630–637 (2013).
Wood, D. J. & Endicott, J. A. Structural insights into the functional diversity of the CDK-cyclin family. Open Biol. 8, 1–26 (2018).
Christensen, C. L. et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26, 909–922 (2014).
Sanso, M. et al. Cdk9 and H2Bub1 signal to Clr6-CII/Rpd3S to suppress aberrant antisense transcription. Nucleic Acids Res. 48, 7154–7168 (2020).
Paparidis, N. F., Durvale, M. C. & Canduri, F. The emerging picture of CDK9/P-TEFb: more than 20 years of advances since PITALRE. Mol. Biosyst. 13, 246–276 (2017).
Zhang, L., Fried, F. B., Guo, H. & Friedman, A. D. Cyclin-dependent kinase phosphorylation of RUNX1/AML1 on 3 sites increases transactivation potency and stimulates cell proliferation. Blood 111, 1193–1200 (2008).
Cummings, J. et al. Alzheimer’s disease drug development pipeline: 2023. Alzheimers Dement. 9, e12385 (2023).
Games, D. et al. Alzheimer-type neuropathology in transgenic mice overexpressing V717f beta-amyloid precursor protein. Nature 373, 523–527 (1995).
Schmitz, C. et al. Hippocampal neuron loss exceeds amyloid plaque load in a transgenic mouse model of Alzheimer’s disease. Am. J. Pathol. 164, 1495–1502 (2004).
Liachko, N. F. et al. CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann. Neurol. 74, 39–52 (2013).
Shackelford, R. E. et al. The Ataxia telangiectasia gene product is required for oxidative stress-induced G1 and G2 checkpoint function in human fibroblasts. J. Biol. Chem. 276, 21951–21959 (2001).
Hoglinger, G. U. et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc. Natl Acad. Sci. USA 104, 3585–3590 (2007).
Sun, X. et al. PANoptosis: mechanisms, biology, and role in disease. Immunol. Rev. 321, 246–262 (2024).
Teo, T. et al. An overview of CDK3 in cancer: clinical significance and pharmacological implications. Pharmacol. Res. 180, 1–15 (2022).
Herrup, K. Post-mitotic role of the cell cycle machinery. Curr. Opin. Cell Biol. 25, 711–716 (2013).
Betz, U. A. K., Voßhenrich, C. A. J., Rajewsky, K. & Müller, W. Bypass of lethality with mosaic mice generated by Cre–loxP-mediated recombination. Curr. Biol. 6, 1307–1316 (1996).
Yuan, J. et al. Two conserved epigenetic regulators prevent healthy ageing. Nature 579, 118–122 (2020).
Caccamo, A. et al. Necroptosis activation in Alzheimer’s disease. Nat. Neurosci. 20, 1236–1246 (2017).
Acknowledgements
We gratefully acknowledge technical support from the Core Facility of Biomedical Sciences and the Laboratory Animal Center at Xiamen University. We extend our deepest appreciation to all colleagues and collaborators whose contributions were instrumental in advancing this research, particularly in elucidating CDK3’s role in AD pathogenesis, developing mouse models, and discovering potent CDK3 inhibitors. The completion of this decade-long project, from conceptualization to paper finalization, reflects the collective perseverance of everyone involved. This work was supported by the National Nature Science Foundation of China (grants 82530048, U23A20430, 81925010, 91849205, and 92149303 to J. Zhang and grants 82101242 and 32371505 to K.Z.), the National Key Research and Development Program of China (grant 2021YFA1101402 to J. Zhang), the China postdoctoral science foundation (grant 2020M682094 to K.Z.), the US National Institutes of Health (grants R01NS120922 and R01AG069912 to K.H.), and the Pennsylvania Department of State (grant 4100087331 to K.H.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
K.Z. and L. Zi contributed equally to this work; J. Zhang conceptualized the study; K.Z., L. Zi, X.S., and Y.S. prepared and maintained the mice; L. Zi conducted MRI imaging on mice; Q.L. and C.H. performed the screening of small-molecule drugs; Y.L. and X.L. performed hESC culturing and generation of cerebral organoids; K.Z. and L.Zi designed and performed morphological analysis and biochemical assays; Y.X., Haojie Li and Shu Wang constructed related plasmids; X.S., Siqi Wang, M.W., and Y.C. conducted electrophysiological recordings. Shuzhong Wang conducted the APEX2-based labeling experiment; B.W., S.C., and L. Zhong reanalyzed the data obtained from the public database; K.Z., L.Zi, J.L., and W.X. conducted data visualization; K.Z., L. Zi, Y.S., X.S., and Y.X. performed the behavior tests; K.Z. and B.W. analyzed single-nucleus sequencing data; K.Z., L.Zi, K.H., and J. Zhang wrote the paper; Y.L., Q.L., L.L., K.H., L. Zhong and J. Zhou discussed and gave suggestions that shaped the paper structure, refined experimental designs and improved the text.; Huifang Li administrated the project; J. Zhang, K.Z., and K.H. supervised the project and acquired funding. All authors reviewed and gave final approval to the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The comparative analysis revealed a premature mutation at Trp-187 in mouse Cdk3, which contributes to enhanced instability and markedly elevated expression levels in AD mouse models (APP/PS1) and AD-associated mouse cell lines relative to wild-type controls.
(a) The region containing a single base mutation of Trp-187 to premature mutant residues in CDK3 (in humans) and Cdk3-ps (in mice) is delineated by a rounded box in both mouse and human samples within the basal segment. (b) The CDK3 proteins derived from mice and humans were compared and the premature mutant points (Trp-187 to Term, termed W187*) are marked in the red box. (c-d) The mRNA level of Cdk3-ps was assessed both N2a cells and N2a-APP-695 cells (N2a cell line expressing human APP695 with the Swedish double mutation), as well as WT and APP/PS1 mouse cortical lysates. (e-f) Western blot analysis of CDK3 and CDK3(aa1-186) degradation rate after CHX treatment (200 μM). Quantification of protein levels are shown in o. Three parallel experiments were performed. (n = 3 cases for N2a and N2a-APP-695 respectively in c. Sample sizes in d: Cdk2, n = 12 for both WT and APP/PS1; Cdk3-ps, n = 48 (WT) and n = 36 (APP/PS1). Data represented mean ± SEM. p < 0.05 indicates significance between the two indicated groups. c, d: two-way ANOVA with Tukey post hoc test.).
Extended Data Fig. 2 The construction and verification of overexpression of human-derived CDK3 (hCDK3) in CCTG mice.
(a) A Schematic diagram of construction of hCDK3-cOE mice with selective conditional overexpression by the CRISPR-CAS9 strategy, where the CDK3 CDS fragment is fused with a HA tag. (b) The efficacy of hCDK3 overexpression was evaluated in the cortex and CA1 region of CCTG mice through immunofluorescence analysis using antibodies against HA and NeuN. Scale bar: 50 μm. (c) The efficacy of hCDK3 overexpression was examined in different brain regions of CCTG mice by Western blotting using antibodies against the HA peptide and CDK3. (d) The flowchart of LC-MS/MS analysis following the enrichment of overexpressed hCDK3 in the hippocampus of CCTG mice. (e-f) LC-MS/MS spectrum illustrating the modification level of the Thr160 residue, a key regulatory site in hippocampal CDK3, which positively correlates with its kinase activity.
Extended Data Fig. 3 CDK3-overexpression aggravates the cognitive impairments and neuronal death in APP/PS1 mice.
(a) Schematic diagrams are shown to illustrate the mating strategy and stereotactic injection strategy. (b) Western blotting experiments confirmed the successful overexpression of CDK3 in the APP/PS1 mice via an injection-based approach. (c-e) The Morris water maze test was performed on four groups of mice. c: The raining curves over six days, d: The time spent in the target quadrant, e: The number of target crossings on the test day. (f) The contextual fear conditioning tests were conducted on these mice. (g) Immunofluorescence co-labeling of CDK3 and GFP confirmed that CDK3 was predominantly overexpressed in the pyramidal neurons of the CA1 region. (h-m) Immunofluorescent staining of NeuN (h-i), Cleaved caspase 3 (j-k) and BrdU (l-m) in CA1 region (the area of injection) from mouse brain samples of these four groups was performed. Quantifications of staining intensity are shown in i, k and m respectively. (h-m: 5-6 slices from 3 mice, 2 slices per mouse). Scale bar: 50 μm. ROI in h: 0.20 mm × 0.20 mm and ROI in k, m: 0.15 mm × 0.15 mm. (All the mice used in behavior tests are male and 9 months old. In Morris water maze test and fear conditioning test, hCDK3-cOE-GFP AAV: 12 mice, hCDK3-cOE-Cre AAV: 12 mice, APP/PS1;hCDK3-cOE-GFP AAV: 12 mice and APP/PS1;hCDK3-cOE-Cre AAV: 13 mice. Data represent mean ± SEM. p < 0.05 indicates significance between the two indicated groups. d, e, f, i, k, m: one-way ANOVA with Tukey post hoc test.).
Extended Data Fig. 4 The recovery of endogenous full-length Cdk3 by correcting the Cdk3 pre-mature mutation.
(a) Generation of a Cdk3-ps (Term-Try187) point mutation mouse model was achieved using CRISPR/Cas9 technology. (b) The relative mRNA levels of Cdk3-ps were analyzed in the brains of Cdk3-ps(A561G/A561G) mouse (Abbreviated as Cdk3(561DM)) and littermate controls using two primer pairs, namely Cdk3-ps-PP1 and Cdk3-ps-PP2, followed by RT-PCR analysis. (c) Western blotting with a c-terminal antibody against Cdk3 revealed the presence of full-length Cdk3 protein in cortical lysates from mutated mice (Cdk3(561DM)), which was absent from their littermate controls. (d-e) Neuronal morphology was detected after treatment with Aβ in primary neuron cells from WT and Cdk3(561DM) mice. e represents statistical analysis of total branch length. Scale bar: 50 μm. (f) After treatment with Aβ, synaptic markers (Snap25, Synaptophysin and Psd95) were assessed in primary neurons from WT and Cdk3(561DM) mice. (Sample number: n = 8 cases for WT and Cdk3(561DM) respectively in b, n = 4 cases for WT and Cdk3(561DM) respectively in c. n = 21-33 neurons per groups, 107 neurons in all in e. Data represent mean ± SEM. p < 0.05 indicates significance between the two indicated groups. b. e: two-way ANOVA with Tukey post hoc test.).
Extended Data Fig. 5 CDK3 drives Aβ pathology and neuroinflammation in 5xFAD mice.
(a-b) Western blot analysis was performed to determine APP and CDK3 protein levels in brain samples from 5xFAD and CCTG;5xFAD mice (a). Quantification of APP levels are shown in panel b (n = 6 samples from 6 mice per group). (c-e) Immunofluorescent staining for Aβ (Red) was carried out in the hippocampus of brain from 9-month-old 5xFAD;hCDK3-cOE mice with injecting AAV-CaMK2α-GFP (Con AAV) or AAV-CaMK2α-Cre (Cre AAV). The quantification and size of Aβ are shown in d and e respectively (6 slices from 3 mice, 2 slices per mouse). Scale bar: 200 μm. ROI: 0.90 mm × 0.90 mm. (f-h) Immunofluorescent staining for Aβ (Red) was carried out in the hippocampus of brain from 10-month-old 5xFAD and CCTG;5xFAD mice. The quantification and size of Aβ are shown in g and h respectively (10 slices from 5 mice, 2 slices per mouse). Scale bar: 200 μm. ROI: 0.90 mm×0.90 mm. (i-j) Histopathological changes in the brains of 5xFAD and CCTG;5xFAD mice were analyzed using Nissl staining. The quantification of the lateral ventricle area is illustrated in b (6 slices from 3 mice; 2 slices per mouse). Scale bar: 5 mm. (k-l) Histopathological changes in the brains of 5xFAD and 5xFAD;Cdk3(561DM) mice were analyzed using Nissl staining. The quantification of the lateral ventricle area is illustrated in d (6 slices from 3 mice; 2 slices per mouse). Scale bar: 1 mm. (m-o) Immunofluorescent staining of Iba1(Green) and GFAP(Red) was conducted in the CA3 region from mouse brain belonging to 5xFAD and CCTG;5xFAD mice. Quantification of Iba1-positive and GFAP-positive cells are shown in n and o respectively (8 slices from 4 mice; 2 slices per mouse). Scale bar: 100 μm. ROI: 0.30 mm × 0.30 mm. (p-r) Immunofluorescent staining of Iba1(Green) and GFAP(Red) was performed in the cortex of mouse brain samples derived from 5xFAD and 5xFAD;Cdk3(561DM) mice. Quantification of Iba1-positive and GFAP-positive cells are shown in q and r respectively (8 slices from 4 mice; 2 slices per mouse). Scale bar: 100 μm. ROI: 0.30 mm × 0.30 mm. (s-u) Immunofluorescent staining of Iba1(Green) and GFAP(Red) was performed in the CA1 region of mouse brain samples derived from 5xFAD and 5xFAD;Cdk3(561DM) mice. Quantification of Iba1-positive and GFAP-positive cells are shown in t and u respectively (8 slices from 4 mice; 2 slices per mouse). Scale bar: 100 μm. ROI: 0.30 mm × 0.30 mm. (Data represent mean ± SEM. p < 0.05 indicates significance between the two indicated groups. b: two-way ANOVA with Tukey post hoc test. j: one-way ANOVA with Tukey post hoc test. d, e, g, h, l, n, o, q, r, t, u: two-sided unpaired t-test.).
Extended Data Fig. 6 CDK3 overexpression induced increased phosphorylation of Tau at S205/T217/S396 in hippocampus, and restoration of CDK3 exacerbates the lateral ventricle enlargement, astrogliosis, and microgliosis in PS19 mice.
(a-b) Representative immunoblots for total Tau and phosphorylated Tau (T181, S202, S202&S205 (AT-8), T217, S396, S404, S416) are shown. The quantitative analysis of phosphorylated Tau is displayed in b (n = 4 independent hippocampal lysates were prepared and analyzed per group). (c-d) Histopathological changes in the brains of PS19 and PS19;Cdk3(561DM) mice were analyzed using Nissl staining. The quantification of the lateral ventricle area is illustrated in d (8 slices from 4 mice; 2 slices per mouse). Scale bar: 5 mm. (e-g) Immunofluorescent staining of Iba1(Green) and GFAP(Red) was performed in the CA1 region of mouse brain samples derived from PS19 and PS19;Cdk3(561DM) mice. Quantification of Iba1-positive and GFAP-positive cells are shown in f and g respectively (8 slices from 4 mice; 2 slices per mouse). Scale bar: 100 μm. ROI: 0.30 mm × 0.30 mm. (Data represent mean ± SEM. p < 0.05 indicates significance between the two indicated groups. b: two-way ANOVA with Tukey post hoc test. d, f, g: two-sided unpaired t-test.).
Extended Data Fig. 7 Restoration of Cdk3 premature mutation accelerates cell cycle reentry-induced neuronal death (CRND).
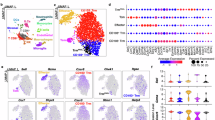
(a-b) The number of genes per nucleus grouped by donor in panel a is as follows: WT: 9,791 nuclei, Cdk3(561DM): 10,797 nuclei, 5xFAD: 9,742 nuclei, 5xFAD;Cdk3(561DM): 9,550 nuclei, PS19: 9,960 nuclei, PS19;Cdk3(561DM): 11,456 nuclei. Additionally, the number of genes per neuronal nucleus grouped by donor in panel b is as follows: WT: 6,664 nuclei, Cdk3(561DM): 7,406 nuclei, 5xFAD: 6,227 nuclei, 5xFAD;Cdk3(561DM): 6,453 nuclei, PS19: 5,959 nuclei, PS19;Cdk3(561DM): 8,941 nuclei. The upper and lower bounds of each violin plot represent the maximum and minimum number of genes detected per nucleus, respectively. In the box plot, the upper and lower whiskers correspond to the upper and lower quartiles, while the hollow central point indicates the mean value. (c) Among the 22 neuronal sub-clusters analyzed for up-regulated genes, cluster 2 contains a total of 960 up-regulated genes. (d) A heatmap displaying the top marker genes for each cluster reveals that Ywhab, Fth1, Ywhah, Prnp, and Rps8 can be considered as marker genes for cluster 2. (e) Scatter plots depicting the expression intensity of marker genes associated with different types of cell death (including apoptosis, necroptosis, ferroptosis and pyroptosis) are presented based on sample source.
Extended Data Fig. 8 BMX330 specifically inhibits the activity of CDK3.
(a) The IC50 values of Dinaciclib and BMX330 for CDK2, CDK3, and CDK9, as well as the IC50 values of THZ1 and BMX330 for CDK7, were determined. Three parallel experiments were performed and the data was measured as a percentage of substrate phosphorylation. IC50 refers to semi-inhibitory concentration. (b-c) Primary neurons derived from WT or hCDK3-OE were cultured and treated with 25 nM BMX330 or vehicle control (Methyl Cellulose, Abbreviate as Met). The kinase activity of CDK3 in these cells was subjected to an in vitro kinase assay measured by Thiophospho ester testing after protein immunoprecipitated using a HA antibody, and 150 ng His-Histone H1 was used as the substrate (b). Quantifications of the kinase assay are shown in c. n = 4 repeats. (d-e) Primary neurons derived from WT or hCDK3 transgenic mice were treated with 25 nM BMX330 for 24 h at DIV10. Double immunofluorescent staining of Map2 and Tuj1 were performed on these cells. The representative staining is shown in d. Quantifications of dendric length are shown in e. n = 15-22 cells per group. Scale bar: 50 μm. (f-g) The effect of BMX330 on CDK2 activity was evaluated using in vitro kinase assays, with Cdk2-IN-22 employed as the positive control. HA-CDK3 was also used as a positive control and subjected to treatment with both inhibitors. The corresponding statistical results are shown in panel g. A total of four parallel experiments were conducted. (h) Upon transfection of 293 T cells with HA-CDK2, the HA tag was enriched, and co-enrichment of endogenous CCNE1 was detected. (i-j) Representative Western blot analysis demonstrates the effect of BMX330 on the phosphorylation level of RB at Ser809 and S811 residues within the context of hCDK3 overexpression, and the statistical results are in j (4 samples per group as shown in i). (Data represent mean ± SEM. p < 0.05 indicates significance between the two indicated groups. c: one-way ANOVA with Tukey post hoc test. e, j, g: two-way ANOVA with Tukey post hoc test.)
Extended Data Fig. 9 CDK3 inhibitor BMX330 alleviates cognitive impairments and neuronal death in hCDK3-overexpressed mice.
(a-b) The pharmacokinetic profile in brain tissue 24 hours after intracranial administration of BMX330. The detailed procedure and the sampling time point are illustrated in a. (c-f) The effects of intracerebroventricular injection of BMX330 or Methyl cellulose (a kind of non-ionic water-soluble polymer, the solvent for BMX330) were evaluated on CDK3-mediated hippocampal atrophy. The workflow for the experiment is outlined in c. Representative pictures of hippocampal sections stained with Cleaved caspase 3 and BrdU are shown in d. Quantifications of Cleaved caspase 3- and BrdU- positive cell number are shown in e and f (6 sections from 6 mice, 1 section per mouse). Scale bar: 200 μm. ROI in d: 0.45 mm × 0.45 mm. (g-k) The effects of intracerebroventricular injection of BMX330 or Methyl cellulose on mouse behaviors were evaluated in CCTG mice. The workflow for the experiment is outlined in g. The Morris water maze test (h-i) and fear conditioning tests (j-k) were performed on these mice. h: The raining curves over six days of training; i: the time spent in the target quadrant on the test day; The contextual (j) and cued (k) fear conditioning tests of the above mice. (All the mice used in behavior tests are male and 3.5-4.5 months old. Methyl cellulose: 10 mice and BMX330: 10 mice. Data represent mean ± SEM. p < 0.05 indicates significance between the two indicated groups. e, f: one-way ANOVA with Tukey post hoc test. i, j, k: two-sided unpaired t-test.).
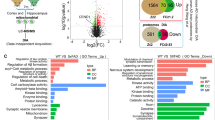
Extended Data Fig. 10 GO and KEGG analysis of multi-omics indicates that CDK3 is involved in the regulation of synaptic function, metabolic process, neuron-death and the cell cycle.
(a) GO and KEGG analysis of CDK3 interactor. (b) GO and KEGG analysis of differentially expressed proteins in the proteome with or without CDK3 overexpression. (c) GO and KEGG analysis of differentially expressed proteins in the phosphoproteome with or without CDK3 overexpression. (Hypergeometric test for enrichment. Yellow boxes denote metabolism-associated GO terms and KEGG pathways; red boxes denote synapse-associated GO terms and KEGG pathways; green boxes denote neuron death-associated GO terms and KEGG pathways; pale pink boxes denote Cell cycle-associated GO terms and KEGG pathways; purple boxes denote neurodegenerative disease-associated GO terms and KEGG pathways. Three biological repetitions were performed for each group in three omics.).
Supplementary information
Supplementary Information
Supplementary Figs. 1–3.
Supplementary Table 1
Gene sets, cycle, and cell death and assessing cellular states at the single-cell level.
Supplementary Table 2
CDK3-associated interactome.
Supplementary Table 3
CDK3-associated proteome.
Supplementary Table 4
CDK3-associated phosphorylated proteome.
Supplementary Table 5
APEX2-CDK3 interactome.
Source data
Source Data Figs. 1–8 and Extended Data Figs. 1–10
Unprocessed blots or gels.
Source Data Figs. 1–8 and Extended Data Figs. 1–10
Unprocessed statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhuang, K., Zi, L., Su, X. et al. CDK3 induces neuronal death and brain atrophy in Alzheimer’s disease. Nat Aging 6, 200–220 (2026). https://doi.org/10.1038/s43587-025-01026-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-01026-6