Abstract
Aging and DNA damage increase the risk of chronic inflammation and autoimmunity, yet the molecular underpinnings remain unclear. In this study, we uncover a DNA damage-driven mechanism in macrophages that triggers immune autoreactivity. Here, using Er1Lyz2/− mice with a macrophage-specific DNA repair defect in ERCC1−XPF, we demonstrate that monocyte-derived macrophages accumulate DNA damage, activate the immune system, drive polyclonal T cell responses and generate antinuclear autoantibodies. Proteomic and immunopeptidomic analyses reveal a distinct major histocompatibility complex class II (MHC-II) antigen repertoire enriched in nuclear and ribosomal peptides, relying on autophagy for nuclear cargo delivery to MHC-II. Aged macrophages exhibit a similar lysosomal cargo profile, linking autophagy-driven nuclear antigen presentation to immune activation. Notably, inhibiting autophagy in Er1Lyz2/− mice suppresses autoimmune features, pinpointing autophagy-facilitated nuclear antigen processing as a central driver of age-related autoimmunity. These findings establish DNA damage-induced autophagy in macrophages as a pivotal mechanism linking aging to autoimmunity, unveiling potential therapeutic targets to mitigate age-related immune dysregulation.
Similar content being viewed by others
Main
Aging is characterized by the accumulation of DNA damage, a hallmark that fuels chronic inflammation, accelerates immunosenescence and drives a spectrum of age-related pathologies1,2,3,4,5,6. Persistent DNA lesions and impaired repair mechanisms activate inflammatory pathways, including type I interferon signaling through cytoplasmic nucleic acid release and the activation of the senescence-associated secretory phenotype (SASP)1,7,8,9,10,11. Recent studies reveal that genotoxic stress stimulates adaptive immunity by generating peptides presented on human leukocyte antigen class I (HLA-I) molecules, highlighting the intricate interplay between DNA damage and immune responses12.
Macrophages have a critical role in the immune system as sentinels, antigen-presenting cells (APCs) and modulators of immunity13,14. Aging impairs macrophage or monocyte function through reduced clearance of apoptotic cells, debris and pathogens15,16, increased MHC expression17, skewed polarization18 and chronic cytokine secretion19. In autoimmune responses, macrophages can contribute to the accumulation and presentation of immunogenic antigens. Identifying these peptides is essential for the development of targeted therapies, such as peptide-based vaccines or immunomodulatory agents, to selectively suppress autoimmune responses without broadly suppressing the immune system20,21.
Autophagy, a fundamental cellular degradation and recycling process, maintains homeostasis by clearing damaged organelles, misfolded proteins and intracellular debris, particularly in response to genotoxic stress22,23. Additionally, autophagy intersects with antigen presentation pathways, playing a pivotal role in MHC-II-mediated antigen presentation24,25. Dysregulation of autophagy-mediated antigen processing is implicated in the pathogenesis of autoimmunity, underscoring its therapeutic potential. Despite its importance, the precise composition of autophagy-derived antigens and their impact on immune activation remain largely undefined26,27. In the present study, we reveal how DNA damage in macrophages drives immune activation using Er1Lyz2/− mice, a model with myeloid-specific DNA repair defects. ERCC1−XPF is a heterodimeric, structure-specific endonuclease complex required for lesion excision in nucleotide excision repair (NER)28,29. It is also known to participate in DNA double-strand break (DSB) repair, DNA interstrand-crosslink (ICL) repair, base excision repair (BER), telomere maintenance and for the processing of various alternative DNA structures that hinder replication and transcription events30,31,32,33. Er1Lyz2/− mice exhibit autoimmune symptoms, driven by MHC-II presentation of nuclear and ribosomal peptides. Aged macrophages show similar increase in antigen presentation, linking these changes to the DNA damage response (DDR). Notably, inhibiting autophagy reduces immune activation, highlighting autophagy as a key driver of nuclear antigen processing and a promising therapeutic target for age-related autoimmune diseases.
Results
Ercc1 ablation in bone marrow-derived macrophages drives spontaneous immune system activation and autoantibody production in mice
To study how persistent DNA damage in monocyte-derived macrophages affects the immune system response, we studied mice with an engineered Ercc1 defect in the myeloid cell lineage (Lyz2-cre+;Ercc1fl/−, hereafter referred to as Er1Lyz2/− mice). By 8 months of age, Er1Lyz2/− mice develop a systemic inflammatory response and an increase in tissue infiltrates compared to their wild-type counterparts—that is, Lyz2-cre−;Ercc1fl/+ mice34. Hematoxylin and eosin (H&E) staining revealed chronic inflammation in multiple intraparenchymal areas of the kidneys isolated from Er1Lyz2/− mice, including around the pelvicalyceal system. The inflammatory infiltrates consisted primarily of small lymphocytes, large activated lymphocytes and numerous plasma cells, indicative of an active immune response (Fig. 1a). Immunofluorescence analysis of kidney cryosections showed an increased deposition of complement protein C3 and immune complexes according to IgM and IgA staining (Extended Data Fig. 1a). These proteins are commonly localized in the kidneys of patients with autoimmune glomerulonephritis and mouse models. Despite this, skin preparations of Er1Lyz2/− mice displayed no inflammatory foci or other alterations according to H&E analysis, and there were no apparent changes in the thickness of paws or ankles (Extended Data Fig. 1b,c). Notably, periodic acid−Schiff (PAS) staining in kidneys and an albumin ELISA argued against the presence of tubular atrophy or fibrosis of the kidney parenchyma or proteinuria in Er1Lyz2/− mice. PAS stain, however, additionally revealed the expanse of the mesangium of 30−40% of the glomeruli in Er1Lyz2/− kidneys, indicating glomerular damage (Extended Data Fig. 1d,e). Supporting this, an indirect immunofluorescence assay using mouse embryonic fibroblasts (MEFs) as a substrate demonstrated the presence of autoantibodies in the sera of 8-month-old Er1Lyz2/− mice. The increase in autoantibody production was similar to that observed in naturally aged 24-month-old wild-type mice (Extended Data Fig. 1f). Antibody staining patterns included homogenous or speckled nuclear and cytoplasmic patterns (Extended Data Fig. 1g). Antinuclear antibody enzyme-linked immunoabsorbent assay (ANA-ELISA) confirmed the presence of antinuclear autoantibodies in the sera of Er1Lyz2/− mice (Fig. 1b). Adoptive transfer of sera derived from 8-month-old wild-type or Er1Lyz2/− mice to young wild-type recipients and analysis of kidney cryosections resulted in an increase in the localization of IgM immune complexes in the glomeruli of mice receiving Er1Lyz2/− sera when they were compared to the recipients of wild-type sera (Extended Data Fig. 2a). Histological analysis of wild-type spleens showed normal morphological features, with a normal ratio of white to red pulp. Er1Lyz2/− spleens, however, showed marked hyperplasia of the white pulp, with foci of plasmacytic cells at the white−red pulp border. Spleen weights complemented these results, with Er1Lyz2/− spleens weighing more than their wild-type controls, pointing to an increase in cellularity (Extended Data Fig. 2b,c). Staining of splenocytes for CD19 (B cell maker) and CD138 (plasma cell marker) revealed increased plasma cell percentages and numbers in the secondary lymphoid organs and the bone marrow and further supported the increased antibody production in 8-month-old Er1Lyz2/− mice (Fig. 1c and Extended Data Fig. 2d,e). This aligns with findings in aged C57BL/6 mice, which spontaneously developed antinuclear autoantibodies driven by MHC-II antigen presentation to CD4+ T cells, accompanied by an immune complex deposition in the kidneys of old mice35. Flow cytometry analysis of splenocytes from Er1Lyz2/− and wild-type mice showed increased numbers of additional cell populations that regulate autoimmunity: CD11b+ myeloid cells, CD4+ cells, CD8+ cells, NK1.1+CD3− (natural killer) cells and regulatory CD4+ T cells in Er1Lyz2/− spleens but not granulocyte−monocyte progenitors in Er1Lyz2/− bone marrow (Fig. 1d−g and Extended Data Figs. 2f−i and 3a). Splenic macrophages (CD11b+F4/80+SinglecF−) and CD11b+Ly6G− cells of monocytic origin from Er1Lyz2/− mice exhibited higher percentages of cells expressing MHC-II antigen presentation and CD86 co-stimulatory molecules, respectively, along with increased protein expression levels of both markers (Fig. 1h and Extended Data Fig. 3b,c). Accordingly, flow cytometry analysis of cells from spleens and lymph nodes stained for the CD4 glycoprotein, the CD44 adhesion molecule responsible for T cell migration and the L-selectin/CD62L naive T cell marker revealed a gradual increase in the percentages and numbers of memory CD4+ T cells (CD44hiCD62Llo) in 8−10-month-old Er1Lyz2/−mice36,37 (Fig. 1i and Extended Data Fig. 3d,e). Conversely, a decrease in the percentage of naive CD4+ T cells (CD44loCD62Lhi) was observed, arguing for a polyclonal CD4+ T cell expansion in Er1Lyz2/− mice (Extended Data Fig. 3f). This resembled the T cell expansion observed in 24-month-old wild-type mice (Fig. 1i and Extended Data Fig. 3g). To assess T cell function, CD4+ T cells from 10-month-old mice were stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin for 4−6 h, which activates protein kinase C and induces calcium signaling, important for T cell activation and cytokine production38. Stimulation of CD4+ T cells from 10-month-old Er1Lyz2/− mice with PMA and ionomycin led to increased interferon-γ (IFNγ) secretion, indicating a more robust T helper 1 (TH1) immune response (Fig. 1j). Correspondingly, a higher percentage of CD4+ T cells expressing T-bet, the master regulator of TH1 differentiation, was observed in the lymph nodes of these mice compared to controls (Fig. 1k and Extended Data Fig. 3h). Along with their activated profile and increased numbers, FOXP3− T cells from Er1Lyz2/− mice increased expression of the inhibitory receptor programmed cell death protein 1 (PD-1) (Fig. 1l) and the proliferation marker Ki-67 (Extended Data Fig. 3I), indicating prolonged exposure to activation signals. Interestingly, adoptive transfer of CD4+ T cells isolated from 10-month-old wild-type and Er1Lyz2/− mice to NOD scid gamma (NSG) hosts led to a higher frequency of T cell infiltration in the kidneys of mice receiving Er1Lyz2/− T cells, as suggested by the inflammatory foci observed during H&E analysis and CD4+ T cell numbers measured by flow cytometry analysis of kidney Percoll fractions (Extended Data Fig. 4a−c).
a, H&E stain of kidneys isolated from 8-month-old wild-type (WT) and Er1Lyz2/− mice (n = 7−8, P = 0.0040). The magnification in each image is indicated, and black arrows point to large activated lymphocytes and plasma cells (inflammation score: 0: no/focal, 1: moderate, 2: intense). b, Antinuclear autoantibody (ANA) detection in the sera of 8−10-month-old WT and Er1Lyz2/− mice by ELISA (n = 8−10, P = 0.0449). Autoantibody patterns of 8-month-old WT and Er1Lyz2/− and 24-month-old WT (aged) mice are shown in Extended Data Fig. 1f,g. c−f,Flow cytometry analysis of 8-month-old WT and Er1Lyz2/− splenocytes stained for plasma cell (CD19, CD138), monocyte-derived cell (CD11b+Ly6G−), CD4+ T cell (CD4) and regulatory T cell (CD4, CD25, FOXP3) markers. All bar plots show the total number of CD19+CD138+ plasma cells (n = 7, P = 0.0201). Myeloid cells (n = 4−5, P= 0.04) (c), CD4+ cells (n = 9−10, P = 0.0469) (d), regulatory T (Treg) cells (n = 3, P = 0.0483) (e) and natural killer (f) and CD8+ T (g) cells (n = 4, P = 0.0004 and P = 0.0395). Representative scatter plots and gating strategies are shown in Extended Data Fig. 2d,f–i. h,Flow cytometry analysis of activated macrophages in spleens of 8-month-old mice using CD11b and Ly6G for gating in cells of monocytic origin (gating strategy in Extended Data Fig. 2f) and MHC-II and CD86 as antigen presentation markers. Representative scatter plots (top) as well as bar plots (bottom) depict the percentage of MHC-II+CD86+ macrophages. MFIs are shown in Extended Data Fig. 3c (n = 10, P = 0.0014). i, Graphs showing the percentage of memory CD4+ T cells in the spleens of 2-month-old, 6-month-old and 10-month-old WT and Er1Lyz2/− mice as well as 24-month-old (aged) WT mice, quantified by flow cytometry analysis. Representative scatter plots for the activation status of T cells isolated from 10-month-old mice are included in the same figure, and representative plots for cells isolated from 2-month-old and 24-month-old mice are shown in Extended Data Fig. 3g (n = 3−7, P = 0.0311 and P < 0.0001). j, ELISA analysis for the detection of IFNγ in the supernatants of PMA and ionomycin-stimulated CD4+ T cells (n = 3, P = 0.0124). k, Bar plot displaying the expression levels of T-bet (MFI) in CD4+ T cells in 8-month-old WT and Er1Lyz2/− spleens. A representative histogram plot can be found in Extended Data Fig. 3h (n = 4−5, P = 0.0182). l, Histogram showing the MFI of PD-1 inhibitory marker in the FOXP3−CD44+ population of WT and Er1Lyz2/− CD4+ T cells in the spleen, at 10 months of age (n = 4, P = 0.0063). m, Adoptive transfer of WT or Er1Lyz2/− BMDMs in young WT mice for a time period of 8−12 weeks, every 2 weeks (experimental scheme shown in top legt). The percentage of activated (CD44hiCD62Llo) CD4+ cells in the inguinal lymph nodes (LN) and the levels of antinuclear autoantibodies in the sera of WT mice receiving WT or Er1Lyz2/− BMDMs are plotted (bottom left and right, respectively). Plasma cells (CD19+CD138+) in the spleens of WT mice receiving WT or Er1Lyz2/− BMDMs are shown in Extended Data Fig. 4e (n = 5−8, P = 0.0374 and P = 0.0320). Representative scatter plots are shown in Extended Data Fig. 4d,e. Error bars indicate s.e.m. among replicates. *P ≤ 0.05 and **P ≤ 0.01 (two-tailed Student’s t-test). NS, not significant.
To determine whether Er1Lyz2/− macrophages could trigger the immune activation observed in Er1Lyz2/− mice, we performed a bone marrow-derived macrophage (BMDM) adoptive transfer experiment. In this experiment, wild-type and Er1Lyz2/− BMDMs, differentiated in vitro, were intravenously administered into young wild-type mice every 2 weeks for 8−12 weeks. Notably, wild-type mice receiving Er1Lyz2/− macrophages showed a higher percentage of memory CD4+ T cells (CD44hiCD62Llo) in their lymph nodes and plasma cells (CD19+CD138+) in their spleens, accompanied by an increase in sera autoantibodies compared to mice receiving wild-type macrophages (Fig. 1m and Extended Data Fig. 4d,e). These results indicate that Ercc1 ablation in BMDMs induces spontaneous immune activation and autoantibody production in mice, further supporting the role of DNA damage in promoting autoimmune and inflammatory responses.
DNA damage in BMDMs drives MHC-II antigen presentation and CD4+ T cell activation
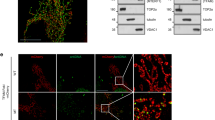
DSBs caused by low-dose chemotherapeutic agents or irradiation upregulate MHC-II via the ATM−NF-κB−IRF1 pathway, a mechanism described in cancer cells and B cells39,40. Similarly, genotoxic stress, such as X-rays or chemotherapeutic drugs, has been predicted to stimulate HLA-I-mediated T cell activation12. DNA-damaging agents such as doxorubicin further link genotoxicity to immunogenicity by inducing senescence and triggering CD8+ T cell responses to neoantigens41. Co-localized foci of DDR markers γH2AX and 53BP1 in Er1Lyz2/− macrophages indicated the accumulation of DNA DSBs, which were partly diminishedby reducing oxidative stress with the use of antioxidants N-acetylcysteine and Mito-TEMPO. This was in line with the higher mean fluorescence intensity (MFI) when we tested for the detection of 8-oxoguanine (8-oxoG) oxidative lesions in Er1Lyz2/− BMDMs (Extended Data Fig. 5a,b). These DNA breaks can activate an ATM-mediated DDR, shown by the increased protein levels of the phosphorylated form of ATM (Extended Data Fig. 5c). To test whether Er1Lyz2/− macrophages harboring DNA damage could directly drive CD4+ T cell responses in an MHC-II-dependent manner, we treated BMDMs with the ATM inhibitor KU-55933 (ref. 42). Er1Lyz2/− BMDMs showed elevated MHC-II levels, a response abrogated by ATM inhibition, demonstrating that ATM activation is critical for DNA damage-driven antigen presentation (Fig. 2a,b). Likewise, treatment of BMDMs with ATR and DNA-PK inhibitors, also key mediators of the DDR, led to a similar decrease in MHC-II levels (Fig. 2b and Extended Data Fig. 5d). DNA damage has been linked to the development of systemic lupus erythematosus (SLE) as a risk factor. For this reason, we considered the role of the DDR in MHC-II antigen presentation in monocyte-origin cells derived from 11-month-old New Zealand Blank (NZB)/New Zealand White (NZW) F1 lupus-prone mice. We performed flow cytometry analysis in splenocytes from lupus-prone and control mice and found increased levels of γ-Η2Α.Χ DNA damage marker in CD11b+Ly6G− monocyte-origin cells (Extended Data Fig. 5e). In addition, the increased antigen presentation capacity of lupus-prone compared to control monocyte-origin cells dropped upon ATM kinase inhibition (Extended Data Fig. 5f). DNA damage thus renders monocytes and macrophages able to initiate innate immune responses in a lupus mouse model, based on immunogenic MHC-II peptide generation. Accordingly, treatment of wild-type BMDMs with etoposide and camptothecin, potent genotoxins that induce DSBs and single-strand breaks (SSBs) through inhibition of topoisomerase II and I, respectively43, activated the DDR. This response, observed 24 h after treatment, resulted in an increase in MHC-II protein levels (Fig. 2c and Extended Data Figs. 5g and 6a). By contrast, stress responses such as tunicamycin-induced endoplasmic reticulum (ER) stress or starvation did not significantly elevate MHC-II levels (Extended Data Fig. 5g). In addition to MHC-II epitope binding to the T cell receptor (TCR), effective T cell activation relies on the cytokine and chemokine milieu provided by APCs44,45,46. Quantitative PCR (qPCR) analysis revealed upregulation of T cell chemoattractant genes in Er1Lyz2/− macrophages (Extended Data Fig. 6b). Building upon findings that DNA damage in brain-resident macrophages induces type I interferon production ex vivo9, we assessed interferon-β (IFNβ) levels in BMDM culture supernatants. Protein concentration and western blot analysis confirmed increased levels of IFNβ in Er1Lyz2/− BMDM supernatants. Additionally, western blotting of BMDM lysates showed increased phosphorylation of STAT1 and upregulation of IRF5, indicating the activation of type I interferon signaling pathways (Extended Data Fig. 6c). MHC-II-mediated antigen presentation is crucial for determining CD4+ T cell reactivity, playing a key role in both peripheral tolerance maintenance and breakdown47,48. To investigate whether Er1Lyz2/− BMDMs could directly activate antigen-experienced CD4+ T cells, we performed a BMDM−T cell co-culture. wild-type or Er1Lyz2/− BMDMs were co-cultured with antigen-experienced CD4+ T cells isolated from 10-month-old Er1Lyz2/− mice, which were more activated in vivo but maintained normative ERCC1 expression levels (Extended Data Fig. 6d). Flow cytometry analysis showed an increase in the percentage of CD4+ T cells expressing CD69, an early activation marker49, when they were co-cultured with Er1Lyz2/− BMDMs compared to wild-type controls (Fig. 2d and Extended Data Fig. 6e). Consistently, IFNγ ELISpot assays, used to measure the effector function of CD4+ T cells in the co-culture, revealed higher IFNγ secretion in the span of 48 h. Although blocking MHC-II or IFNβ levels both led to a reduction in the number of IFNγ spots, MHC-II led to a substantial reduction in the spot numbers, whereas IFNβ blockade led to a smaller but consistent decrease (Fig. 2e). These results confirm the enhanced capacity of Er1Lyz2/− macrophages to activate T cells, linking DNA damage in macrophages to an activated adaptive immune response and implicated antigen presentation and type I interferon in this process. Considering the activation status of macrophages, we hypothesized that MHC-II-mediated antigen presentation would be critical for sustaining CD4+ T cell activation in Er1Lyz2/− mice. To test this, we blocked the MHC-II−TCR interaction in vivo by intraperitoneally administering an anti-MHC-II or an isotype control antibody to 6-month-old wild-type and Er1Lyz2/− mice, which showed a similar percentage of activated CD44hiCD62Llo T cells at this age (Figs. 1i and 2f). Five weeks after treatment, at the point when Er1Lyz2/− untreated mice showed an activated immune system (Fig. 1 and Extended Data Figs. 1−3), there was a reduction in MHC-II surface levels in CD11b+ myeloid and in CD4+ T cell activation percentages in Er1Lyz2/− mice treated with anti-MHC-II compared to the isotype control-treated group. This was evident by the CD44hiCD62Llo population decrease and reduced IFNγ secretion after PMA and ionomycin stimulation. By contrast, no changes in T cell activation were observed in wild-type mice, regardless of anti-MHC-II or isotype control treatment (Fig. 2g,h and Extended Data Fig. 6f,g). Moreover, we reported a drop in the C3 complement protein deposition in the glomeruli of anti-MHC-II-treated Er1Lyz2/− mice when they were compared to their isotype controls (Extended Data Fig. 6h). To further support our data, we next performed a depletion of CD4+ T cells by administration of a neutralizing anti-CD4 antibody to 6-month-old Er1Lyz2/− mice for 5 weeks. This led to a reduction in the inflammatory foci in the kidneys and a decrease in plasma cell numbers and percentages in spleens derived from anti-CD4-treated mice compared to the ones derived from isotype control-treated mice (Fig. 2i,j and Extended Data Fig. 6i).
a, Flow cytometry analysis of WT and Er1Lyz2/− BMDMs and Er1Lyz2/− BMDMs treated or not with an ATM inhibitor (ATMi), stained with the antigen presentation protein MHC-II. The histograms show the surface MHC-II expression levels of BMDMs (MFI). Isotype controls are shown in gray. Gating strategy is indicated in Extended Data Fig. 5d (n = 3, P = 0.0029 and P = 0.0015). b, Western blotting of MHC-II protein levels in whole-cell extracts from WT and Er1Lyz2/− DMSO-treated controls (Ctrl) or ATMi-treated or ATR kinase inhibitor (ATRi)-treated BMDMs. Actin was used as a loading control (n = 4−5, P = 0.0244, P = 0.0441 and P = 0.0133). NEL, normalized expression levels. c, Flow cytometry analysis of the MHC-II expression levels of WT control and WT etoposide (ETO)-treated BMDMs. An overlay of representative histograms is shown, and MFIs are plotted. An isotype control is shown in gray (n = 3, P = 0.0189). d,e, Co-culture of WT or Er1Lyz2/− BMDMs and 10-month-old Er1Lyz2/− CD4+ T cells in a 1:1 or 1:4 ratio (n = 5, P = 0.0316). d, 1:1 ratio. The rectangle gate and the corresponding bar plots mark the percentage of activated CD4+ T cells in culture (CD69+CD4+ cells). e, 1:4 ratio. ELISpot assay measuring the secretion of T-cell-derived IFNγ. Representative images and corresponding bars in the plot show the number of spots per 2 × 105 cells plated. Er1Lyz2/− BMDMs were untreated or treated with anti-IFNβ (aIFNβ) or anti-MHC-II (aMHC-II) neutralizing antibodies (n = 4−5, P = 0.0006, P = 0.0354 and P < 0.0001). f−h, In vivo MHC-II blockade. f, The experimental scheme indicates that mice were treated with an aMHC-II or an isotype control antibody for a total of 6 weeks, receiving intraperitoneal injections once per week. m, months. g, The graph shows the percentage of activated CD4+ T cells (CD44hiCD62Llo) in the spleens of WT and Er1Lyz2/− mice treated with an aMHC-II or an isotype control antibody (n = 5, P = 0.0213 and P = 0.0265). Representative plots are shown in Extended Data Fig. 6f. h, ELISA analysis. The plot depicts the secreted IFNγ protein levels in the PMA and ionomycin-stimulated CD4+ T cells isolated from Er1Lyz2/− mice receiving either an MHC-II-blocking or an isotype control antibody (n = 4−5, P = 0.0447). i, Experimental scheme for a 6-week CD4+ T cell depletion in Er1Lyz2/− mice. Mice received injections once per week intravenously. j, CD4+ T cell depletion experiment. Representative scatter plots and graphs of the plasma cell percentages and numbers in spleens of Er1Lyz2/− mice receiving isotype an isotype control or an anti-CD4 (aCD4) antibody (n = 5−6, P = 0.0020 and P = 0.0357 for percentages and P = 0.0002 and P = 0.0007 for counts). Error bars indicate s.e.m. among replicates.*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (two-tailed Student’s t-test).
DNA damage alters the macrophage antigenic landscape
The MHC-II-mediated generation of CD4+ T cells in Er1Lyz2/− mice prompted us to examine if there are DNA damage-related epitopes driving this phenotype. We characterized the immunopeptidome of wild-type and Er1Lyz2/− BMDMs as well as wild-type macrophages treated with lipopolysaccharide (LPS), a well-known immunostimulant50. LPS induces higher MHC-II protein expression while maintaining a similar DDR in wild-type and LPS-treated BMDMs (Extended Data Fig. 7a,b). After MHC-II peptide immunoprecipitation (Extended Data Fig. 7c) and peptide purification, we performed liquid chromatography−tandem mass spectrometry (LC−MS/MS) (Fig. 3a), identifying 1,237 peptides from wild-type mice, 1,290 peptides from Er1Lyz2/− mice, and 999 peptides from LPS-treated wild-type BMDMs. Of these, 798, 823 and 661 peptides were predicted to be potential MHC-II binders for each condition, respectively.
a, Experimental outline for the preparation and mass spectrometry analysis of MHC-II-bound peptides from WT and Er1Lyz2/− untreated and LPS-treated BMDMs. IP, immunoprecipitation. Created in BioRender. Arvanitaki, E. (2025) https://BioRender.com/m68w132. b, Histogram displaying the range of the peptide amino acid (aa) length. All peptides from all samples (WT, Er1Lyz2/− and LPS-treated WT) were used for this graph. c,Gibbs clustering depicting the predominant MHC-II core amino acids. The peptides that were classified as low or high binders of MHC-II from all samples were included for this analysis. NetMHCIIpan (version 4.3; H-2-IAb and H-2-IAq alleles) was used for the identification of peptides with any binding affinity. d, Volcano plot of differentially presented peptides in WT (downregulated, blue) and Er1Lyz2/− (upregulated, red) cells. log2(fold change) of −6 or +6 represents peptides uniquely identified in the WT or Er1Lyz2/− MHC ligandome. Statistical significance was set at P ≤ 0.05 (ANOVA analysis) (horizontal black dashed line) and peptide enrichment at log2(fold change) ≥ 0.3 (upregulated in Er1Lyz2/−) or log2(fold change) ≤ −0.3 (downregulated in Er1Lyz2/−) (vertical black dashed line). Nuclear and ribosomal as well as uniquely identified proteins are labeled with their corresponding gene symbol. e, Bar charts showing the intracellular origin of peptide antigens significantly enriched in the WT, Er1Lyz2/− and LPS-treated WT samples. WT overrepresented peptides were compared to Er1Lyz2/− ones, whereas Er1Lyz2/− and LPS-treated WT enriched peptide sequences were both compared to WT controls. Bold numbers correspond to the percentages of peptides derived from nuclear proteins. ECM, extracellular matrix. f, Venn diagram for the comparison of Er1Lyz2/− overrepresented peptides with the previously characterized peptides in the IEDB. g, Bubble plot of the Gene Ontology (GO) term enrichment analysis (Cellular Component, Mann−Whitney U-test) of significantly overrepresented Er1Lyz2/− peptides, when compared to WT controls (P ≤ 0.05 and log2(fold change) ≥ 0.3). The dot size shows the total count of genes per annotated pathway, and the blue color scale indicates the statistical significance as per the P value of the enriched pathways. The x axis indicates the fold enrichment derived from pathway analysis. h, IFNγ ELISpot analysis of splenocytes isolated from either 8-month-old WT or Er1Lyz2/− mice and pulsed with peptides derived from the indicated source proteins in the presence of IL-2. Representative images are shown in this figure or in Extended Data Fig. 7f. Er1Lyz2/− unpulsed, IL-2-exposed splenocytes were used as a negative control (red dotted line) (n = 4; exact P value is provided in the Source Data file). Error bars indicate s.e.m. among replicates. *P ≤ 0.05 and **P ≤ 0.01 (two-tailed Student’s t-test).
Most peptides were 14–17 amino acids in length (Fig. 3b), aligning with the most common peptide length presented by MHC-II proteins. Moreover, peptide clustering using GibbsCluster confirmed the presence of the typical MHC-II binding motifs (Fig. 3c). Notably, more presented peptides were enriched or uniquely identified in Er1Lyz2/− BMDMs compared to untreated and LPS-treated wild-type cells, indicating that DNA damage alters the antigenic landscape of macrophages (Fig. 3d, Extended Data Fig. 7d and Supplementary Table 1). These overrepresented peptides predominantly originated from subcellular compartments including the ER (calnexin), ribosomal proteins (RPL30 and RPS19) and, notably, the nucleus (such as histone H1, NOLC1, HNRNPL, EXOSC4 and NME2). By contrast, peptides from wild-type cells were mostly derived from plasma membrane and cytosolic proteins, whereas peptides presented by LPS-treated wild-type cells were from extracellular proteins (MMP14 and FN1) and plasma membrane proteins (ANPEP and MARCKSL1) (Fig. 3e). Comparison of the Er1Lyz2/− enriched peptides with Immune Epitope Database (IEDB) peptides identified 72 epitopes arising under genotoxic stress, including those from histone H1, NOLC1, HNRNPL and NME2 proteins (Fig. 3f). Gene Ontology (Cellular Component) enrichment analysis of the significantly overrepresented Er1Lyz2/− peptides further highlighted an increased abundance of epitopes associated with the nucleus, euchromatin, perinuclear space and ribonucleoprotein complexes (Fig. 3g). Conversely, pathway analysis in the overrepresented LPS-treated wild-type antigens confirmed their distinct origin (Extended Data Fig. 7e).
To assess the predicted immunogenicity of the Er1Lyz2/− overrepresented peptides, we synthesized peptides, isolated splenocytes from 8-month-old wild-type or Er1Lyz2/− mice and performed IFNγ ELISpot assays to restimulate CD4+ T cells. We selected peptides that were uniquely identified or of nuclear origin, as nuclear antigens are crucial for the development of autoimmune responses51. We found that peptides of nuclear origin (H1 and HNRPL) and the ribosomal and nuclear peptide RPL30 elicited a more robust IFNγ T cell response in Er1Lyz2/− splenocytes (Fig. 3h and Extended Data Fig. 7f).
DNA damage in macrophages enhances MHC-II-dependent T cell responses by changing the autophagic cargo of cells
To explore how genotoxic stress shapes the antigen presentation profile in Er1Lyz2/− cells, we focused on the mechanisms that enable self-peptides to be presented by MHC-II molecules. These peptides are typically delivered to MHC-II via two pathways: the endocytosis/phagocytosis pathway, where external materials are engulfed by APCs, and the autophagy-mediated pathway. Both pathways culminate in protein degradation in lysosomes52,53. Given that DNA damage activates autophagy in macrophages34, we examined its role in enhancing immunogenicity. To assess the contribution of autophagy to MHC-II-dependent T cell responses, we inhibited autophagosome formation in Er1Lyz2/− BMDMs by treating them with 3-methyladenine (3-MA)54. When treated macrophages were co-cultured with CD4+ T cells from 8-month-old Er1Lyz2/− mice, we observed a reduction in the number of spots in ELISpot assays (Fig. 4a). This provides direct evidence that autophagy has a crucial role in MHC-II peptide presentation in DNA-damaged macrophages.
a, IFNγ ELISpot analysis of recall assay: Er1Lyz2/− CD4+ T cells isolated from 8-month-old mice were co-cultured with WT, Er1Lyz2/− or Er1Lyz2/− 3-MA-treated BMDMs. Representative images of the wells are shown, and the number of spots per reaction is plotted (n = 4, P = 0.033 and P = 0.0379). b−d, Autophagosome content identification in the U2-OS cell line. b, Experimental scheme of the APEX2−LC3B-based proteomics approach. BP, biotin-phenol. Created in BioRender. Arvanitaki, E. (2025) https://BioRender.com/t43t868. c, Volcano plot of proteins enriched in the autophagosomes of DMSO-treated (downregulated, blue) or ETO-treated (upregulated, red) U2-OS cells. Statistical significance (two-tailed Student’s t-test) was set at P ≤ 0.05 (horizontal black dashed line) and peptide enrichment at log2(fold change) ≥ 0.85 (overrepresented in autophagosomes derived from ETO-treated cells, ≥1.75 fold change) or log2(fold change) ≤ −0.85 (overrepresented in autophagosomes derived from DMSO-treated cells, ≤−1.75 fold change) (vertical black dashed line). Nuclear, ribonucleoprotein complex and ribosomal proteins are labeled with their corresponding gene symbol. d, Bubble plot of the GO term enrichment analysis (Cellular Component, Mann−Whitney U-test) of significantly overrepresented proteins found in autophagosomes isolated from ETO-treated cells (P ≤ 0.05 and log2(fold change) ≥ 0.85). The dot size shows the total count of genes per annotated pathway, and the color scale indicates the statistical significance as per the P value of the enriched pathways. e, Western blot detection of lamin A/C and lamin B1 protein levels in whole-cell extracts from WT and Er1Lyz2/− BMDMs. Actin was used for normalization (n = 3, P = 0.04397 and P = 0.048812). f, Immunofluorescence staining for the detection of cytoplasmic chromatin fragments and histone H1 upon treatment of WT cells with ETO and/or autophagy inhibitor chloroquine and Er1Lyz2/− cells with autophagy inhibitors chloroquine (CQ), 3-MA and BafA1. Ctrl, untreated control. White square boxes indicate areas selected for zoomed-in images per genotype per treatment, displayed on the top left of each image panel. White arrows indicate H1+DAPI+ cytosolic chromatin fragments, and green arrowheads point to the focal accumulation of cytoplasmic H1 species. The graphs show the percentage of H1+DAPI+ structures in the cytoplasm of WT or Er1Lyz2/− cells (graph on the left) and the MFI of histone H1 measured in the cytoplasm of cells (graph on the right) (5−8 fields for cyto-DAPI and three independent optical fields for cyto-H1 were counted from each biological replicate; exact P value is provided in the Source Data file). Error bars indicate s.e.m. among replicates. *P ≤ 0.05 and **P ≤ 0.01 (two-tailed Student’s t-test). Scale bars, 10 μm.
Cells experiencing senescence or DNA damage rely on autophagy to remove excess material55,56,57,58,59,60,61. We hypothesized that DNA damage-induced alterations in autophagosome content could determine the availability of proteins for lysosomal processing and MHC-II loading. To explore this, we employed an in vivo proximity biotinylation approach, combining the APEX2−LC3B system with proteomic analysis62. We constructed U2-OS cells constitutively expressing the APEX2−LC3B construct, cultivated them in SILAC (stable isotope labeling by amino acids in cell culture) media for metabolic labeling and exposed them to etoposide or dimethyl sulfoxide (DMSO). Cells were labeled with ‘medium’ and ‘heavy’ amino acids, respectively, and subjected to biotinylation by addition of biotin-phenol and H2O2, whereas non-biotinylated controls were labeled with ‘light’ amino acids. After treatment, cells were exposed to bafilomycin A1 (BafA1), a vacuolar H+ ATPase (V-ATPase) inhibitor, to accumulate autophagosomes and prevent degradation of their contents63,64.
Autophagosomal cargo was purified by streptavidin pulldown in the presence of proteinase K to prevent contaminants from the outer membrane of autophagosomes, and the protein content was quantified using LC−MS/MS (Fig. 4b). Etoposide treatment led to the enrichment of 128 proteins within autophagosomes, including vesicle-resident proteins such as MAP1LC3B and LAMP1, as well as nuclear-derived proteins such as histones H2A, H2B and H3, LMNA, NME2 and HNRNPL and ribosomal proteins such as FAU, RPL23A and RPL4 (Fig. 4c,d and Supplementary Table 2). These results are consistent with the peptides identified in the Er1Lyz2/− BMDM immunopeptidome, highlighting a substantial overlap in the pathways involved. This suggests that DNA damage-induced changes in the autophagic cargo are pivotal in shaping the antigenic profile of macrophages, contributing to the MHC-II loading of nuclear-derived peptides.
Autophagy drives nuclear homeostasis in Er1 Lyz2/− macrophages
Our findings uncover a pivotal role for autophagy in maintaining nuclear homeostasis under genotoxic stress. Previous studies linked the loss of lamin A/C and lamin B1 to DNA damage, which may lead to the release of nuclear constituents into the cytoplasm, where they can be cleared via autophagy. To test this, we examined the levels of these nuclear lamins in wild-type and Er1Lyz2/− BMDMs. We observed a reduction in both lamin A/C and lamin B1 in Er1Lyz2/− BMDMs (Fig. 4e), suggesting that DNA damage disrupts nuclear integrity, despite the lack of cell cycle arrest (Extended Data Fig. 8a). Interestingly, ATR-mediated but not ATM-mediated signaling seems to be associated with the decrease in lamin B1 levels (Extended Data Fig. 8b). We further investigated the consequences of autophagic inhibition by treating BMDMs with 3-MA, BafA1 and chloroquine, which led to the accumulation of cytoplasmic chromatin structures (DAPI+H1+) and nucleosome protein H1 in Er1Lyz2/− macrophages (Fig. 4f). Similarly, etoposide-treated wild-type cells displayed a phenotype resembling that of Er1Lyz2/− macrophages. No DSBs were induced within a 3-h timeframe by these treatments, as evidenced by the co-localization of γH2AX and 53BP1 markers (Extended Data Fig. 8c). The cGAS−STING pathway, a key sensor of cytosolic DNA, is involved in the removal of DNA through autophagy, as autophagy proteins interact with cGAS−STING to promote clearance of cytosolic double-stranded DNA65,66. We tested the levels of the cytosolic cGAS in wild-type and Er1Lyz2/− BMDMs and found increased levels in untreated Er1Lyz2/− BMDMs compared to wild-type controls. Autophagic induction via starvation in Er1Lyz2/− macrophages, however, reduced cytosolic cGAS levels, suggesting that increased autophagy turnover facilitates the removal of chromatin fragments (Extended Data Fig. 8d). To monitor autophagic flux in these cells, we transfected BMDMs with a fluorescent mCherry−GFP−LC3 plasmid67. In Er1Lyz2/− macrophages, we observed a higher red-to-green fluorescence ratio, indicating increased loss of GFP fluorescence (sensitive to acidic lysosomal environments), whereas red fluorescence was retained in the lysosomes, suggesting an enhanced autophagic flux (Extended Data Fig. 8e). Moreover, lysosomes in Er1Lyz2/− BMDMs were larger and more acidic than in wild-type controls, as shown by flow cytometry analysis with LysoTracker and LysoSensor dyes (Extended Data Fig. 8f,g). Collectively, our data point to an enhanced lysosomal function, likely enabling the more efficient and timely transfer of nuclear material for the generation of antigens in Er1Lyz2/− BMDMs.
To further investigate the role of autophagy in the transport of nuclear material to lysosomes, we performed immunofluorescence studies with antibodies against LAMP-1, p62, histone H1 and lamin B1. Cells were treated with chloroquine to inhibit the degradation of autophagic cargo. We observed co-localization of H1 and lamin B1 proteins with autophagosomes, which were either fusing with or in close proximity to lysosomes. This was quantified by measuring the percentage of cells with triple co-localized foci and the total number of these foci per each individual cell (Fig. 5a,b). Interestingly, treatment of Er1Lyz2/− cells with Dynasore68, a dynamin inhibitor that blocks endocytosis and phagocytosis in macrophages (early endosome antigen EEA1; Extended Data Fig. 9a), did not diminish the levels of H1 or lamin B1 co-localizing with autolysosomes. This enhanced the notion that autophagic vacuoles dispose of dispensable cytoplasmic content originating from within Er1Lyz2/− cells rather than cell debris derived from neighboring macrophages (Fig. 5a,b and Extended Data Fig. 9b). Furthermore, this is supported by the lack of cell death, as measured by flow cytometry analysis of Annexin V/propidium iodide-stained cells (Extended Data Fig. 9c). Finally, we purified and analyzed lysosomes from chloroquine-treated wild-type and Er1Lyz2/− BMDMs. Western blotting confirmed the presence of nuclear proteins such as histone H1, lamin B1, lamin A/C and FAU ubiquitin-like as well as ribosomal protein S30 and the LAMP-1 lysosomal protein (Fig. 5c and Extended Data Fig. 9d). These findings underscore the enhanced nucleophagy and turnover of ribosomal and nuclear proteins in Er1Lyz2/− macrophages, highlighting the crucial role of autophagy in maintaining nuclear integrity on the one hand but enrichment of nuclear antigens in the lysosomes on the other hand. Notably, evidence from immunofluorescence studies in the THP-1 human monocytic cell line upon etoposide treatment and monocyte-derived CD11b+Ly6G− cells isolated from SLE model mice additionally revealed the presence of small cytoplasmic chromatin fragments. These DNA moieties are coated with histone H1 and co-localized with autophagy protein p62 (Fig. 5d,e). Etoposide-induced DNA damage additionally upregulated HLA-DR (MHC-II cell surface receptor) in THP-1 cells, further verifying our hypothesis in human monocytes (Extended Data Fig. 9e).
a,b, Co-localization studies of autophagy (p62), lysosomes (LAMP-1) and histone H1 (a) or lamin B1 (LMNB1) (b) upon chloroquine or chloroquine and Dynasore (Dyn) treatment. Magenta arrowheads point to H1+p62+LAMP-1+ (a) or LMNB1+p62+LAMP-1+ (b) foci. White arrows point to cytoplasmic chromatin fragments. Single-channel or two-channel images and higher magnifications of a are shown in Extended Data Fig. 9b and Supplementary Fig. 2a,b. The two graphs illustrate the percentage of cells with any triple co-localized foci (n = 3 biological replicates, 4−7 optical fields) or the total number of triple co-localized foci per each individual cell counted (n ≥ 144 cells for H1 and n ≥ 162 cells for LMNB1). c, Western blot analysis of lysosomes purified from WT and Er1Lyz2/− BMDMs, after chloroquine treatment for 3 h. Membranes were probed for ribosomal (FAU) and nuclear (H1, LMNB1 and LMNA/C) markers. LAMP-1 was used as a resident protein of lysosomes. The splice point is indicated by a vertical black line. Equal amounts of protein (up to 10 μg) were loaded, and protein levels were normalized according to Ponceau stain. The normalized (Norm.) protein levels are plotted (n = 3−4). d,e, Immunofluorescence staining of THP-1 human monocytes (n = 3, P = 0.0053) (d) and cells of monocytic origin sorted from NZB/NZW F1 mice (SLE) (n = 3 P = 0.0115) (e), stained for DAPI, H1 and p62. Cells were fixed after a 3-h treatment with chloroquine. Magenta arrowheads point to chromatin fragments co-localized with p62. The graphs illustrate the percentage of cells with cytoplasmic chromatin fragments. Error bars indicate s.e.m. among replicates. *P ≤ 0.05, **P ≤ 0.01 and ****P < 0.0001 (two-tailed Student’s t-test). Scale bars, 10 μm. Exact P values are provided in the Source Data.
Autophagy inhibition ameliorates inflammation and T cell activation in Er1 Lyz2/− mice
To further investigate the role of autophagy in modulating immune responses in Er1Lyz2/− mice, we generated double knockout (DKO) mice by simultaneously deleting Ercc1 and Atg5, which is crucial for autophagosome formation, in myeloid cells (Ercc1fl/−;Atg5fl/fl;Lyz2-cre+) (Extended Data Fig. 10a). In comparison to their Er1Lyz2/− littermates, DKO mice exhibited a reduction in inflammatory foci in their kidneys at 8 months of age, accompanied by lower levels of antinuclear autoantibodies in their sera and moderate hyperplasia of the spleen white pulp with plasmacytic cell foci (Fig. 6a,b and Extended Data Fig. 10b). The numbers of CD11b+F4/80+ splenic macrophages and Lin−c-Kit+Sca-1−CD34+CD16/32+ myeloid granulocyte−monocyte progenitor cells in the bone marrow were similar between Er1Lyz2/− and DKO mice (Extended Data Fig. 10c,d). Building on this, we checked for the percentages of activated CD4+ T cells and plasma cells and found them to be decreased in DKO compared to Er1Lyz2/− spleens (Fig. 6c,d and Extended Data Fig. 10e). Assessment of cell viability in wild-type, Er1Lyz2/− and DKO BMDMs exhibited no alterations among genotypes, indicating that neither Ercc1 loss nor autophagy ablation in the absence of Ercc1 induces cell death in BMDMs (Extended Data Fig. 10f). To test whether the lack of Er1Lyz2/− nuclear and ribosomal antigen presentation could induce the mitigation of adaptive immune system activation, we next performed MHC-II peptidomics analysis in DKO BMDMs (Supplementary Table 3). We selected the potential MHC-II binders and searched for the protein names of the peptides uniquely identified in the case of Er1Lyz2/− immunopeptidomes when they were compared to wild-type control peptidomes. Notably, no peptides derived from H1.1, RPL30, EXOSC4 and ATP5ME were identified in any of the wild-type or DKO MHC-II ligandomes. Likewise, most of the rest of the Er1Lyz2/− peptides (corresponding to 103 out of 120 proteins) were not in common with the DKO overrepresented peptides. We then checked for the subcellular origin of the DKO overrepresented peptides and traced them back to mainly plasma membrane, cytosolic and extracellular matrix proteins, whereas 8% of the peptides consisted of nuclear proteins where approximately 30% of them were overrepresented in Er1Lyz2/− cells (Fig. 6e). Considering that DNA damage in BMDMs triggers cytoplasmic DNA sensing coupled with a type I interferon response, we checked for the cytoplasmic cGAS levels also in DKO BMDMs, which were increased in comparison to wild-type cells (Extended Data Fig. 10g). DKO BMDM−CD4+ T cell co-culture data well aligned with these results, eliciting a dampened IFNγ production compared to T cells cultured with Er1Lyz2/− BMDMs (Extended Data Fig. 10h). Furthermore, ELISpot assays using splenocytes from 8-month-old mice demonstrated a decline in the recall response of antigen-specific CD4+ T cells isolated from DKO mice (Fig. 6f), indicating a critical role for autophagy in systemic inflammation and autoimmune feature appearance in this model. Interestingly, when we purified lysosomes from DKO BMDMs, there was a reduction in the levels of histone H1 and nuclear lamins B1 and A/C compared to lysosomes from Er1Lyz2/− BMDMs. These findings suggest that autophagy is essential for the presentation of immunogenic nuclear and ribosomal antigens (Fig. 6g).
a, H&E staining in kidney sections derived from 8-month-old WT, Ercc1Lyz2/− and DKO mice. Magnification is indicated. Black arrows point to inflammatory foci. The inflammation score is shown, where 0: no/focal and 1: moderate. The red dotted line represents the mean inflammation score in the kidneys of Er1Lyz2/− mice (n = 8, P = 0.0313). b, ANA-ELISA for the detection of antinuclear autoantibodies in the sera of 8-month-old DKO mice. The red dotted line indicates the average titer of antinuclear antibodies in the Er1Lyz2/− sera (n = 6, P = 0.0449). c, Percentages of CD44hiCD62Llo activated CD4+ T cells in the spleens of 8-month-old WT, Er1Lyz2/− and DKO mice (n = 5−6, P = 0.0009 and P= 0.0404). d, Percentages (left) and numbers (right) of plasma cells in the spleens of 8-month-old WT, Er1Lyz2/− and DKO mice (n = 5−6). e, Bar chart depicting the subcellular origin of the peptides overrepresented in DKO BMDMs compared to WT BMDMs. PEAKS software was used. The bold number corresponds to the percentage of peptides derived from nuclear proteins f, IFNγ ELISpot assay. Splenocytes were isolated from 8-month-old Ercc1Lyz2/−, and DKO mice were pulsed with immunogenic peptides. Representative images are shown, and the number of spots is plotted (n = 3−4, P = 0.005838 and P = 0.000025). g, Western blot analysis of H1 and lamin A/C nuclear proteins present in lysosomal extracts derived from Ercc1Lyz2/− and DKO BMDMs. Equal amounts of protein (up to 10 μg) were loaded, and protein levels were normalized according to Ponceau stain. The plots show the fold change (FC) of the levels of the indicated proteins in DKO versus Ercc1Lyz2/− lysosomes (n = 3) (the latter indicated with the red dotted line). Error bars indicate s.e.m. among replicates. *P ≤ 0.05 and **P ≤ 0.01 (two-tailed Student’s t-test). Exact P values are provided in the Source Data.
Aged macrophages exhibit DNA damage, autophagy-driven lysosomal changes and enhanced antigen presentation
The lysosomal proteome of senescent cells changes significantly, contributing to the lysosome-derived SASP69. In addition to the exocytosis of extracellular matrix components, we hypothesized that lysosomes from senescent or aged cells might be enriched with proteins also found in the lysosomes of BMDMs accumulating DNA damage (such as in Er1Lyz2/− cells), potentially influencing the immunopeptidome. To test this, we analyzed the lysosomal proteome of senescent SK-MEL-103 cells induced by palbociciclib70. We found that approximately 25% of the Er1Lyz2/− MHC-II-presented peptides were shared with the lysosomal cargo of senescent cells, including nuclear proteins H1 and NME2 and ribosomal proteins such as RPL30 and RPL35A (Fig. 7a). Next, we investigated whether macrophages from aged mice display similar traits of senescence and genotoxic stress, potentially altering their lysosomal content and immunopeptidome. We examined different monocyte and macrophage subtypes from both young (2 months) and aged (24 months) mice, including BMDMs (Ly6G−CD11b+CD115+), cells of monocytic origin (Ly6G−CD11b+) and peritoneal macrophages staining positive for marker F4/80 (thioglycolate-elicited macrophages (TEMs)) (Supplementary Fig. 1a−c). Aged BMDMs exhibited elevated levels of γH2X compared to their young counterparts. Similarly, more splenic macrophages and TEMs from aged mice were positive for γH2AX (Fig. 7bi(iii)). Flow cytometry analysis showed that MHC-II levels were increased across all three aged macrophage subtypes, indicating enhanced antigen presentation (Supplementary Fig. 1d). Notably, a greater proportion of aged TEMs tested positive for the senescence-associated β-galactosidase (SA-β-gal) marker, similar to Er1Lyz2/− BMDMs34 (Fig. 7c). Aged TEMs also showed lamin A/C loss (Supplementary Fig. 1e). Immunofluorescence analysis revealed a higher percentage of cells with chromatin fragments and higher co-localization of histone H1, p62 and LAMP-1 in aged TEMs (Fig. 7d). These results suggest that DNA damage in aged macrophages leads to profound alterations in their autophagosomal and lysosomal cargo, similar to senescent cells, thereby contributing to higher antigen presentation. Finally, we investigated whether nuclear proteins such as histone H1 and lamin A/C were transported more frequently in lysosomes of aged TEMs and found that they contained higher levels of both proteins. This increase was reduced with 3-MA, implicating macroautophagy in this process (Fig. 7e). Thus, both senescent cells and aged macrophages exhibit significant changes in their lysosomal proteomes, with increase in antigen presentation and, likely, alterations in their immunopeptidome. These changes, driven by DNA damage and macroautophagy, highlight the potential for lysosomes to influence immune responses during aging.
a, Venn diagram showing the comparative analysis of the lysosomal proteome derived from palbociciclib-induced senescent SK-MEL-103 cells (blue) and of the MHC-II-bound peptidome of Er1Lyz2/− BMDMs (green). The numbers indicate the number of proteins in each dataset. b, γH2AX levels in aged monocytes or macrophages. Isotype (Iso.) controls are shown in gray. (i) Flow cytometry analysis of cells isolated from bone marrow of young (2 month (m)) and aged (24 month) mice, stained for Ly6G, CD11b, CD115 surface and γΗ2ΑΧ DNA damage markers. Representative histogram overlay of γH2AX in bone marrow monocytes (Ly6G−CD11b+CD115+ cells) and representative bar plot of the γH2AX MFI (n = 3−4). (ii) Flow cytometry analysis of splenocytes isolated from young and aged mice, stained for Ly6G, CD11b and γΗ2ΑΧ. A representative histogram overlay of γH2AX levels in cells of monocytic origin (Ly6G−CD11b+ cells), with the black bisector gate indicating the γH2AX+ population. The bar plot indicates the corresponding percentages of γH2AX+ cells (n = 4). (iii) Immunofluorescence staining of TEMs isolated from young and aged peritonea using a γH2AX antibody. Representative images and quantification of the percentage of γH2AX+ cells are shown (5−10 optical fields were counted) (n = 6). c, Representative images of the SA-β-gal assay using young and aged TEMs. The percentage of β-gal+ cells is plotted (three independent optical fields, and over 360 cells were counted per biological replicate) (n = 4, P < 0.0001). d, Representative images of the immunofluorescence detection of H1, p62 and LAMP-1 in young and aged TEMs. The white arrow points to a cytoplasmic structure stained positive for DAPI and all three proteins. The graphs depict the percentage of cells with chromatin fragments (top, DAPI+H1+) and of cells with H1+p62+LAMP-1+ foci (4−7 independent optical fields, and n > 67 cells were counted, respectively) (n = 3−5) P = 0.0093 and P = 0.0133. Single-channel images of d are shown in Supplementary Fig. 2c. e, Western blot analysis of lysosomal preparations derived from young and aged TEMs, treated or not with 3-MA, as indicated. All were treated with chloroquine for the inhibition of lysosomal cargo degradation. LAMP-1 protein levels are shown as a positive control and Ponceau stain as a loading control. In each case, 7 μg of protein was loaded. A quantitative analysis is presented in the bar chart (n = 3−5). The splice point is indicated by a vertical black line. Error bars indicate s.e.m. among replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (two-tailed Student’s t-test). Scale bars, 10 μm. Exact P values are provided in the Source Data.
Discussion
Our study reveals how DNA damage in macrophages links genotoxic stress to autoimmune disorders and identifies autophagy as key in nuclear and ribosomal antigen processing. This mirrors findings in DNA repair-deficient conditions such as xeroderma pigmentosum, where impaired genome maintenance predisposes individuals to autoimmune conditions such as type 1 diabetes and SLE71. Similarly, type 1 diabetes has been associated with Cockayne syndrome, driven by transcription-coupled repair defects that impair DNA damage removal from actively transcribed genes72,73. Individuals with Cockayne syndrome exhibit increased inflammation74. Consistently, exposure to genotoxins can trigger autoimmune responses75,76,77,78. In line, individuals with SLE exhibit higher DNA damage levels compared to healthy individuals, and several proteins related to genome maintenance have polymorphisms79,80,81. Pharmacological targeting of ATR in B cells of patients with active SLE disease attenuates antibody production82. Accordingly, monocyte-derived cells isolated from NZB/NZW F1 mouse spleens exhibited higher levels of γH2AX and MHC-II depending on the DDR protein ATM.
Native DNA turns immunogenic when modified or mislocalized—a shift evident in SLE, where the presence of anti-DNA antibodies is a hallmark. In our study, Er1Lyz2/− mice developed antinuclear autoantibodies, kidney inflammation and complement deposition in the glomeruli, hyperplasia of the spleen white pulp and activation of adaptive immunity. Kidney targeting of immune complexes may be a result of the accumulation of tissue-external nuclear antigens and their corresponding antinuclear antibodies in the kidney filtration barrier or the outcome of MHC-II antigen presentation from BMDMs in the glomerular microvasculature and resident macrophages83,84. DNA damage or repair defects lead to cytoplasmic accumulation of single-stranded DNA10, double-stranded DNA9 or telomeric DNA85, activating a type I interferon response. Cells clear cytoplasmic DNA via various mechanisms including autophagy to maintain homeostasis and prevent immune activation. In addition, autophagy is implicated as a critical mechanism in the defense against pathogens86. Besides its role in suppressing inflammatory responses, macroautophagy is also known to contribute to the development of intracellular MHC-II antigens87,88. Genetic depletion of autophagy in APCs—that is, dendritic cells or macrophages—results in impaired epitope presentation via MHC-II and epitope-specific CD4+ T cell responses89,90,91,92,93. In vivo, dendritic cell-specific Atg5-deficient mice have abrogated autoimmune disease appearance upon myelin oligodendrocyte glycoprotein (MOG) immunization and fail to mount effective TH1 responses in the presence of a viral challenge, due to defective processing and presentation of MHC-II antigens89,90,93. In Er1Lyz2/− cells and genotoxin-treated macrophages and human monocytes, DNA damage boosts MHC-II antigen presentation, displaying nuclear self-peptides, as revealed by MHC-II peptidome analysis. This underscores the role of autophagy in clearing nuclear proteins for antigen presentation. The enrichment of nuclear proteins and ribosomal protein FAU in lysosomes from Er1Lyz2/− BMDMs supports this hypothesis. Our data align with previous reports that different pathologies result in the formation of unique MHC-II peptidomes94,95. Myeloid cell-specific DNA repair and autophagy-deficient mice (Ercc1fl/−;Atg5fl/fl;Lyz2-cre+, DKO) showed improved autoimmune symptoms, reduced antigen-driven CD4+ T cell responses and reversed nuclear and ribosomal MHC-II epitope presentation and nuclear protein enrichment in lysosomes (Fig. 7f). This supports that the mechanism of action of hydroxychloroquine and chloroquine—drugs commonly used to treat rheumatic diseases—involves reducing the ability of APCs to present immunogenic peptides to CD4+ T cells.96. Consistently, monocytic-origin cells isolated from NZB/NZW F1 mice had a higher percentage of chromatin fragments in their cytoplasm that were co-localized with p62. Conversely, enhancing autophagy via starvation in Er1Lyz2/− BMDMs lowered the levels of cytoplasmic double-stranded DNA sensor cGAS, indicating the role of autophagy in preventing toxic buildup and dampening antiviral responses to cytoplasmic DNA. Over time, however, this protective mechanism may shift, driving chronic antigen load and heightened T cell responses, ultimately contributing to the development of immunological memory with age. Aged monocytes in mice and humans show elevated MHC-II levels, signaling higher antigen loads and increased autoimmune risk with age17. In 24-month-old macrophages, this rise in antigen presentation, along with increased DNA damage, was evident. We propose that increased nuclear component trafficking to lysosomes via macroautophagy drives heightened MHC-II presentation of nuclear peptides. Together with the observed DNA damage in aged monocytes, this suggests that aging fosters the generation of immunogenic epitopes in macrophages. SASP components secreted during aging significantly contribute to macrophage immunogenic potential and likely also autoimmune responses. In this work, besides the aged macrophage secretome, we suggest that senescent features also determine their lysosomal proteome and MHC-II peptidome. Studies in animal models have shown that chloroquine extends the lifespan in Caenorhabditis elegans, mice and rats while reducing systemic inflammation97,98,99. Similarly, 3-MA alleviates colitis and inflammation in aged mice100. In this respect, boosting DNA repair capacity101, developing DNA damage-centered peptide vaccines102, leveraging exosome-based therapies to remove cytoplasmic nucleic acids9,10,85 or inhibiting autophagy in age-related autoimmune disorders is promising and could enhance immune tolerance and reduce autoimmune risk.
Methods
Animals
Wild-type and Er1Lyz2/− mice were generated as previously described by intercrossing Lyz2-cre (C57BL/6 background), Ercc1fl/fl (FVB background) and Ercc1+/− (C57BL/6 background) mice. This breeding strategy produced wild-type mice carrying a floxed and a wild-type Ercc1 allele (Ercc1fl/+) and Er1Lyz2/− mice carrying one floxed and one knockout allele in conjunction with the Lyz2-cre transgene (Lyz2-cre;Ercc1fl/−). Similarly, Atg5fl/fl mice (C57BL/6) were used to achieve the conditional knocking out of both Atg5 and Ercc1 genes in the same background. Eleven-month-old male F1 mice exhibiting lupus-like disease resulted from the cross of NZB × NZW mice. Mice were housed in a specific pathogen-free facility at the Institute of Molecular Biology and Biotechnology-Foundation for Research and Technology (IMBB-FORTH) where the light/dark cycle (12 h) and temperature were controlled. Mice were fed a normal chow diet and were provided water ad libitum. This work received ethical approval by an independent animal ethics committee at IMBB-FORTH. All relevant ethical guidelines for the work with animals were adhered to during this study. For the duration of all in vivo experiments, mice were monitored daily.
Isolation of mouse sera and adoptive transfer in young hosts
Mice were initially anesthetized using ketamine/xylazine. For the acquisition of serum, the blood was centrifuged twice at 10,000g for 10 min at 4 °C. The supernatant was kept, and pellets were discarded. For the adoptive transfer of mouse sera to young hosts, sera from 10-month-old wild-type and Er1Lyz2/− mice were diluted 1:3 in 1× PBS and injected intravenously once per week for a total of 5 weeks.
Primary BMDM culture, lentiviral transfection and treatments
Bone marrow was harvested from the tibias and femurs of mice, and precursor cells were differentiated in DMEM supplemented with 10% FBS, antibiotics (50 μg ml−1 streptomycin, 50 U ml−1 penicillin from Sigma-Aldrich, 2 mM l−1 glutamine from Gibco) and 30% L929 conditioned media for 6 days. On the seventh day, 30% L929 media were replaced with fresh DMEM containing 10% FBS, antibiotics and 10% L929. All treatments were performed on the seventh day of differentiation. In more detail, etoposide (ETO; Sigma-Aldrich, E1383) was added at the concentration of 25 μM for 1 h, and cells were recovered for 24 h before MHC-II flow cytometry analysis. Inhibitors targeting ATM kinase signaling (10 μM ATMi; KU 60019; Sigma-Aldrich, 531978), ATR kinase signaling (10 μΜ ATRi; Millipore, 189299) and DNAPK kinase signaling (2.5 μΜ; NU7441; STEMCELL Technologies, 74082) were added for a 6-h duration. N-acetylcysteine (Sigma-Aldrich, A9165) and Mito-TEMPO (Sigma-Aldrich, SML0737) were added at the concentration of 1 mM and 20 μM, respectively, for 24 h. Autophagy inhibitors 3-MA (Sigma-Aldrich, 189490), 10 mM, and chloroquine (Sanofi Aventis), 50 μM, were added for a duration of 3 h. For the inhibition of endocytosis, cells were treated with 80 μM Dynasore (Sigma-Aldrich, 324410) for a total of 3 h, including a 2-h chloroquine treatment in the case of autolysosome−nuclear protein co-localization studies. LPS-induced activation of BMDMs was carried out at a concentration of 100 ng ml−1 for a 16-h timeframe. For the determination of the cells’ autophagic flux, FUW mCherry−GFP−LC3 lentivirus (Addgene, plasmid no. 110060; http://n2t.net/addgene:110060; RRID: Addgene_110060) production was performed in HEK293T cells with helper plasmids psPAX2 and pMD2.G. The supernatant was collected 72 h later, filtered using a 45-μm filter and precipitated with polyethylene glycol before immediate use or storage at −80 °C until use. Viral transfection of BMDMs with the mCherry−GFP–LC3 plasmid was performed in 10% L929 conditioned media on day 6 of differentiation. Cells were fixed 48 h later, permeabilized with 0.1% Triton in 1× PBS and stained with DAPI for the detection of nuclei.
THP-1 monocyte cell line
Cells were purchased from the American Type Culture Collection (no. TIB-202) and cultured in RPMI 140 (Gibco). Approximately 2 × 105 cells were treated with 25 μM ETO for 1 h or left untreated and recovered in fresh RPMI medium for 24 h before flow cytometry analysis or 4% formaldehyde fixation for immunofluorescence staining. Cells analyzed with immunofluorescence were treated with chloroquine for 3 h beforehand.
Primary CD4+ T cell isolation and co-culture with BMDMs
CD4+ T cells were isolated from spleens of mice. Single-cell suspensions of splenocytes were obtained by mushing spleens in 40-μm strainers, collecting cell pellets by centrifugation at 300g for 5 min at room temperature, removing red blood cells (RBCs) by resuspending the pellets in RBC lysis buffer for 2 min at room temperature and, finally, centrifuging again at 300g for 5 min at room temperature. MACS MicroBeads (CD4 L3T4; Miltenyi Biotec, 130-117-043) were used for positive selection of the desired cell population, as per the manufacturer’s instructions. For the T cell−BMDM co-culture, isolated BMDMs were seeded on a 96-well plate with CD4+ T cells in a ratio of 1:1 in the presence of 0.5 μg ml−1 CD28 (Invitrogen, 16-0281-86), in RPMI 140 medium (Gibco).
Adoptive transfer experiments
BMDMs were differentiated as described, and 5 × 106 cells were injected intravenously at the timepoints indicated. CD4+ T cells were isolated as described, and 2 × 106 cells were injected intravenously once per week for a total of 8 weeks in young NSG hosts.
In vivo MHC-II blockade
Intraperitoneal injections of 70 μg of anti-MHC-II (I-A/I-E, M5/114 monoclonal antibody by Bio X Cell) blocking or isotype control antibodies were administered weekly, for 4−6 weeks.
In vivo depletion of CD4+ T cells
Intravenous injections of 150 μg of anti-CD4 (Bio X Cell) blocking or isotype control antibodies were administered weekly, for 6 weeks.
Immunofluorescence of cells and indirect detection of antinuclear antibodies
BMDMs, TEMs, THP-1 monocytes, CD11b+Ly6G− monocytes from NZB/NZW F1 mice or MEFs were fixed in 1× PBS/4% formaldehyde for 10 min at room temperature. Cells were washed three times in 1× PBS, blocked and permeabilized using 1% BSA and 0.1% Triton X-100 in 1× PBS. Primary antibodies or mouse sera were incubated in 1% BSA/0.1% Triton X-100/PBS for 1 h at room temperature or overnight at 4 °C. Afterwards, cells were washed three times with 0.1% Triton/PBS, and fluorochrome-conjugated secondary antibodies were added in 1% BSA/0.1% Triton X-100/PBS for 1 h at room temperature, followed by three more washes in 0.1% Triton X-100/1× PBS. Mounting was done with 80% glycerol, and samples were imaged with a Leica SP8 confocal microscope.
8-oxoG staining preparation
Cells were fixed in methanol, on ice, for 10 min and washed three times with 1× PBS. After fixation, the coverslips were air dried and incubated in 0.05 N HCl for 5 min, on ice, washed three times in 1× PBS and then incubated in a 100 μg ml−1 RNase A (Macherey-Nagel, 740397), 150 mM NaCl and 15 mM sodium citrate solution for 1 h, at 37 °C. A 1× PBS wash was performed for 3 min, followed by sequential ethanol dehydration steps: 35%, 50% and 75% ethanol for 3 min. Then, 0.15 N NaOH in 70% ethanol was added for 4 min; two PBS washes were performed; and cells were fixed with 4% formaldehyde in 70% ethanol for 2 min. The fixation buffer was exchanged with 50% and then 35% ethanol, and cells were again washed with 1× PBS for 2 min and treated with 5 μg ml−1 proteinase K in Tris-EDTA buffer for 5−10 min, at 37 °C. After a PBS wash, blocking and primary antibody incubation was performed as described before. An 8-oxoG antibody (Millipore, MAB3560) was used, at a 1:100 concentration, overnight.
Immunofluorescence of kidney cryosections
Kidneys were fresh frozen in OCT compound and stored at −80 °C until further analysis. Kidneys were sliced using a Leica CM1850 UV cryostat (7 μm). Tissue sections were fixed in 4% formaldehyde for 15 min, washed three times with 1× PBS and blocked with 1% BSA/0.1% Triton X-100/PBS for 1 h at room temperature. A similar protocol as the one for immunofluorescence of fixed cells was followed. Kidneys were stained for IgM (dilution 1:500; Invitrogen, A21042), IgA (dilution 1:800; BioLegend, 407001) or C3 (dilution 1:300; Invitrogen, PA5-21349).
Paraffin preparations for tissue histological analysis
Skins, kidneys and spleens were dissected from mice, fixed overnight in 4% formaldehyde, washed three times with 1× PBS and then embedded in paraffin blocks. Tissue sections were used for H&E or PAS staining.
Flow cytometry analysis and antibodies
For surface protein staining, cells were stained with fluorochrome-conjugated antibodies diluted in staining buffer (1× PBS/5% FBS or 1× HBSS/5% FBS) for 20 min on ice, using the concentrations indicated by the manufacturer. Cells were washed by staining buffer and centrifuged at 300g for 5 min at 4 °C. For the staining of intracellular proteins, True-Nuclear Transcription Factor Buffer Set (BioLegend, 424401) was used, and cells were centrifuged at 400g for 5 min at room temperature, after fixation. Staining for the detection of granulocyte−monocyte progenitors in the bone marrow was as follows. Bone marrow was collected by flushing the femur, and cells were incubated with RBC buffer for the removal of erythrocytes. Then, 1 × 106 cells were stained with Pacific Blue anti-mouse lineage antibody cocktail (1:10), PE anti-mouse CD34 (1:50), FITC anti-mouse c-Kit (1:100), APC anti-mouse CD16/32 (1:50) and PerCP anti-mouse SCA-1 antibody (1:100) for 3 h at 4 °C, before a PBS/5% FBS wash and flow cytometry analysis. Antibodies and isotype controls were purchased from BioLegend and Proteintech: anti-CD19 (BioLegend, 152410; clone 1D3/CD19), anti-CD138 (BioLegend, 142503; clone 281-2), anti-CD11b (BioLegend, 101212; clone M1I70), anti-CD4 (BioLegend, 100406, 100412 and 100432; clone GK1.5), anti-CD25 (BioLegend, 102012; clone PC61), anti-FOXP3 (Proteintech, PE-65089; clone 3G3), anti-MHC-II (BioLegend, 107606, 107636 and 107631; clone M5/114.15.2), anti-CD86 (BioLegend, 105026; clone GL-1), anti-Ly6G (BioLegend, 127654 and 127607; clone 1A8), anti-CD62L (BioLegend, 104412; clone MEL-14), anti-CD44 (BioLegend, 103036; clone IM7), anti-T-bet (BioLegend, 644812; clone 4B10), anti-PD-1 (BioLegend, 135214; clone 29F.1A12), anti-CD69 (BioLegend, 104507; clone H1.2F3), anti-F4/80 (BioLegend, 123110; clone BM8), anti-CD115 (BioLegend, 135512; clone AFS98), anti-mouse Lineage Cocktail with Isotype Ctrl (BioLegend, 133305; clones 17A2, RB6-8C5, RA3-6B2, Ter119 and M1/70), anti-mouse CD34 antibody (BioLegend, 119307; clone MEC14.7), anti-mouse Ly-6A/E (Sca-1) (BioLegend, 108123; clone D7), anti-mouse CD16/32 (BioLegend, 101325; clone 93), anti-mouse c-Kit (BioLegend, 105815; clone 2B8), anti-mouse CD170 (Siglec-F) antibody (BioLegend, 155523; clone S17007L), anti-mouse NK-1.1 (BioLegend, 108705; clone PK316), anti-mouse CD3a (Proteintech, PE-65060), anti-mouse FoxP3 (BioLegend, 126409; clone MF-14), i-mouse CD8a (BioLegend, 100712; clone 53-6.7), PerCP rat IgG2a (BioLegend, 400529; clone RTK2758) and FITC rat IgG2a (Proteintech, FITC-65209; clone 2A3) were used as isotype control antibodies. Lysosomal dyes were purchased from Thermo Fisher Scientific: LysoTracker Red DND-99 (L7528) and LysoSensor Green DND-189 (L7535). For Annexin V/propidium iodide staining in BMDMs, the FITC Annexin V Apoptosis Detection Kit (BD Pharmingen, 556547) was used. Cell analysis was eventually performed in a FACSCanto II flow cytometer or a FACSCalibur (BD Biosciences), and data analysis was performed using FlowJo software (Tree Star). The gating strategies were as follows: forward scatter/side scatter (FSC/SSC) for live cell selection and debris removal; forward scatter area/side scatter area (FSC-A/SSC-A) for the subsequent removal of cell aggregates; and then fluorophore-conjugated specific antibodies for the next gates.
FACS and treatment of NZB/NZW F1 monocytes
Spleens were obtained from NZB/NZW F1 mice, and single-cell suspensions were either cryopreserved and thawed for analysis or directly stained with PE anti-mouse Ly6G (1:200) and APC anti-mouse CD11b (1:100) for 20 min, at 4 °C, in 1× PBS/5% FBS and 2 mM EDTA. Cell sorting was performed in a FACSAria III flow cytometer, and monocytic-origin cells were identified as Ly6G−CD11b+. Cells were seeded on poly-l-lysine-coated coverslips, in 48-well plates (105 cells per well), and cultured for 24 h in RPMI 1640 and recombinant M-CSF (250 ng ml−1 working concentration; PeproTech, 315-02). Cells were treated with chloroquine for 3 h prior to fixation. The splenocytes that were not separated through cell sorting were seeded on 48-well plates (105 cells per well), in RPMI 1640 and recombinant M-CSF, and treated with an ATM inhibitor for a duration of 16 h. Cells were collected and stained with DAPI for dead cell exclusion, CD11b and Ly6G for monocyte labeling and MHC-II antigen presentation protein.
qPCR
Quantitative qPCR was performed using a CFX Duet Real-Time PCR system device (Bio-Rad), and data were analyzed as previously described34. The Hprt1 (hypoxanthine phosphoribosyltransferase 1) gene was used for normalization. Primers: Ccl2: forward: TGATCCCAATGAGTAGGCTGGAG, reverse: ATGTCTGGACCCATTCCTTCTTG; Ccl7: forward: TCCCTGGGAAGCTGTTATCTTC, reverse: TGGAGTTGGGGTTTTCATGTC; Ccl24: forward: AATTCCAGAAAACCGAGTGG, reverse: TGGCCCCTTTAGAAGGCTGG; Cxcl1: forward: CCACACTCAAGAATGGTCGC, reverse: GTTGTCAGAAGCCAGCGTTC; Cxcl5: forward: TGCCCCTTCCTCAGTCATAG, reverse: GGATCCAGACAGACCTCCTTC; Cxcl10: forward: ATGACGGGCCAGTGAGAATG, reverse: CATCGTGGCAATGATCTCAACA.
Lysosome purification
At least 30 × 106 BMDMs or TEMs pooled from four mice were collected for the isolation of lysosomes using the Lysosome Isolation Kit (Abcam, ab234047). Lysosomes were then lysed using RIPA, and their protein content was detected through immunoblotting analysis.
Western blot analysis and antibodies
Cells or lysosomes were lysed with RIPA buffer, containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40 and 0.1% sodium dodecyl sulfate and protease and phosphatase inhibitors. For IFNβ detection from BMDMs, culture supernatants from the same amount of cells were concentrated using Amicon Ultra Centrifugal Filter, 10-kDa molecular weight cutoff (Merck Millipore, UFC901024). The concentrated supernatants were mixed with equal volumes of 2× Laemmli and boiled at 80 °C for 10 min before being loaded into the gel for SDS−PAGE. For cell and lysosome lysates, protein concentration was determined using Bradford protein assay, and equal amounts of protein were loaded (50−80 μg for cells and 5−8 μg for lysosomes) for SDS−PAGE. Equal parts of concentrated supernatant proteins were loaded. Proteins were transferred to nitrocellulose membranes (Amersham Hybond), blocked using 5% skim milk diluted in 1× PBS with 0.1% Tween 20 (PBS-T) for 1 h and probed with antibodies. β-tubulin or actin was used for the normalization in the case of cell lysates and supernatants and Ponceau staining for the normalization in the case of lysosomal lysates. An ECL (Thermo Fisher Scientific and Amersham) development was performed, and results were imaged using ImageBlot (Bio-Rad). Quantification was performed using Fiji (ImageJ). Antibodies against the following proteins were used: MHC-II (Bio X Cell, clone M5/114; western blot: 1:800), ERCC1 (Santa Cruz Biotechnology, clone D-10; western blot: 1:500), Ki-67 (Cell Signaling Technology, 9129S, clone D3B5; FACS: 1:500), γH2AX (Millipore, 05-636; immunofluorescence: 1:12,000), 53BP1 (NB100-304; immunofluorescence: 1:200), IFNβ (Cell Signaling Technology, 97450; immunofluorescence: 1:500, western blot: 1:1,000), pSTAT1 (Cell Signaling Technology, 9167; western blot: 1:250) and STAT1 (Cell Signaling Technology, 14994; western blot: 1:500), β-tubulin (Abcam, ab6046; western blot: 1:1,000), IRF5 (Proteintech, 10547-1-AP; western blot: 1:500), actin (Cytoskeleton, BK037; western blot: 1:5,000), H1 (Santa Cruz Biotechnology, sc-8030; immunofluorescence: 1:50, western blot: 1:200), LAMN A/C (Proteintech, 10298-1-AP; western blot: 1:2,000), lamin B1 (Abcam, ab16048; immunofluorescence: 1:500, western blot: 1:1,000), cGAS (Proteintech, 26416-1-AP; immunofluorescence: 1:200), EEA1 (Proteintech, 28347-1-AP; western blot: 1:200), LAMP1 (Santa Cruz Biotechnology, sc-19992; immunofluorescence: 1:100), LAMP1 (Developmental Studies Hybridoma Bank; western blot: 1:200), p62 (Abnova; immunofluorescence: 1:1,000), p62 (Cell Signaling Technology; immunofluorescence: 1:500), GAPDH (Abcam, ab8245; western blot: 1:2,000), FAU (Proteintech, 13581-1-AP; western blot: 1:200), ATG5 (Proteintech, 10181-2-AP; western blot: 1:1,000) and γH2AX (Cell Signaling Technology; immunofluorescence: 1:500, FACS: 1:500).
MHC-II immunoprecipitation and peptidomics analysis
Approximately 3 × 108 BMDMs, derived from a pool of isolated cells from four mice of the same genotype, were used per each MHC-II immunoprecipitation sample. Cells were initially lysed using a buffer containing 0.5% NP-40, 50 mM Tris (pH 8.0), 150 mM NaCl and protease inhibitors. After rotating the lysates for 1 h at 4 °C, the samples were centrifuged at 2,000g for 10 min at 4 °C, and supernatants were again centrifuged at 51,200g for 50 min at 4 °C. Native MHC-II−peptide complexes were purified using InVivoMAb anti-mouse MHC-II (I-A/I-E, M5/114 monoclonal antibody by Bio X Cell) along with Protein G Sepharose beads (Millipore). Immunopeptides were purified using Sep-Pak tC18 columns containing 100 mg of sorbent (Waters Corporation). The elution of peptides from the tC18 sorbent was conducted with 32% acetonitrile (ACN) in 0.1% trifluoroacetic acid (TFA). Eluates were volume reduced using a vacuum evaporator until almost all liquid was evaporated. The peptides were then resolved with 2% ACN in 0.5% TFA and stored at −80 °C until further analysis103.
LC−MS/MS and quantitative analysis
An LC−MS/MS analysis was performed on a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific) online coupled to an UItiMate 3000 RSLC nano-HPLC (Dionex/Thermo Fisher Scientific). The peptides were automatically injected and loaded onto a C18 trap column (300-µm inner diameter × 5 mm, Acclaim PepMap100 C18, 5 µm, 100 Å, LC Packings; Thermo Fisher Scientific) at a 30 µl min−1 flow rate prior to performing C18 reversed-phase chromatography on the analytical column (nanoEase MZ HSS T3 Column, 100 Å, 1.8 µm, 75 µm × 250 mm; Waters Corporation) at a 250 nl min−1 flow rate in a 95-min nonlinear ACN gradient from 3% to 40% in 0.1% formic acid. Profile precursor spectra from 300 m/z to 1,650 m/z were recorded at 60,000 resolution with an automatic gain control (AGC) target of 3 × 106 and a maximum injection time of 100 ms. The 15 most abundant peptide ions of charges 1 to 4 were selected from the mass spectrometry scan and fragmented using higher-energy collisional dissociation (HCD) with a normalized collision energy of 28, an isolation window of 1.6 m/z and a dynamic exclusion of 15 s. MS/MS spectra were recorded at a resolution of 30,000 with an AGC target of 1 × 105 and a maximum injection time of 100 ms. Proteome Discoverer 2.5 software (version 2.5.0.400; Thermo Fisher Scientific) was used for peptide and protein identification via a database search (Sequest HT search engine) against the SwissProt murine database (release 2020_02; 17,070 sequences). Furthermore, the workflow for the identification of the immunopeptidome included the INFERYS rescoring node104. The database search was performed with an unspecified peptide cleavage. The precursor mass tolerance was 10 ppm, and the fragment mass tolerance was 0.02 Da. The carbamidomethylation of cysteine was set as static modification. Dynamic modifications included the deamidation of asparagine and glutamine, the oxidation of methionine and a combination of methionine loss with acetylation on the protein N terminus. Peptide spectrum matches (PSMs) and peptides were validated with the Percolator algorithm105. Only the top-scoring hits for each spectrum were accepted with a false discovery rate (FDR) < 1% (high confidence).
The mass spectrometry data for Fig. 6 were acquired in DDA-PASEF mode on a timsTOF Ultra 2 mass spectrometer (Bruker). Peptides were loaded on Evotips (one Evotip for each injection). They were placed in an Evosep autosampler until analysis. The 40 samples per day whisper method employing a 27-min gradient with solvents A (0.1% formic acid, water) and B (0.1% formic acid, MeCN) was chosen, and a 15-cm column (PepSep C18, 1.9-µm beads, 75-µm inner diameter) was used for separation of peptides. Column was heated to 50 °C. The DDA-PASEF method (‘MHC class II’) covered a mass range from 100 m/z to 1,700 m/z and a mobility range from 0.64 to 1.45 1/K0. A duty cycle of 100% was achieved by setting the ramp time and accumulation time to 100 ms each. Collision energy for 0.6 1/K0 was set to 20 and for 1.6 1/K0 to 59. Estimated cycle time was 0.6 s.
PEAKS Studio 12.5 software (Bioinformatics Solutions) was employed for peptide identification using the DeepNovo algorithm for de novo peptide sequencing against the SwissProt Mouse database (release 2020_02; 17,061 entries). Mass spectrometry data were searched without enzymatic specificity constraints, with precursor mass error tolerances set to 10.00 ppm and 20.00 ppm for different experiments, and fragment mass error tolerance was maintained at 0.02 Da. Peptide length was constrained between 6 and 30 amino acids to accommodate the broader range typical for MHC-II immunopeptidomics applications. Variable post-translational modifications included N-terminal acetylation (+42.01 Da), asparagine and glutamine deamidation (+0.98 Da) and methionine oxidation (+15.99 Da), with a maximum of three variable modifications permitted per peptide. PSMs were filtered using stringent criteria including DeepNovo confidence scores ≥70.00% and PSM significance thresholds of −log10(P) ≥ 20.0, with confident amino acid assignment requiring ≥2.00% threshold. Label-free quantification was performed using identification-directed quantification with feature intensity thresholds ranging from 300 to 100,000. Ion mobility tolerance was set to 0.05 (1/K0) where applicable. Data refinement included mass correction and chimera association algorithms to improve spectral quality. No normalization methods were applied to preserve the inherent biological variance in the quantitative data.
ELISAs
Co-culture supernatants were collected after two centrifugations: 300g, 5 min, room temperature, for the removal of cells and 2,000g, 15 min, 4 °C, for the removal of cell debris. Protease inhibitors were added, and the samples were stored at −80 °C until use. Mouse IFNγ protein levels were quantified using Mouse IFNγ ELISA MAX Deluxe Set (BioLegend, 430804). Antinuclear autoantibodies were quantified using Mouse Anti-Nuclear Antigens (ANA/ENA) Ig (total (A + G + M)) ELISA Kit (Alpha Diagnostics International, 5210). Albumin levels in the urine were detected using Mouse Albumin ELISA Kit (Bethyl Laboratories, E99-134).
IFNγ ELISpot assay
ELISpot assays were performed according to the manufacturer’s instructions (Mabtech, 3321-4APT-2). In total, 200.000 splenocytes were stimulated for 48 h in the presence of 3 μg of the indicated synthesized peptide (Macrogen) and 30 U ml−1 recombinant interleukin-2 (rIL-2; PeproTech, 0717108). rIL-2-stimulated splenocytes derived from Er1Lyz2/− mice were used as a negative control. For the T cell−macrophage co-cultures, T cells and BMDMs were mixed in a 1:4 ratio for 48 h, in the presence of 0.5 μg ml−1 anti-CD28 and 30 U ml−1 rIL-2. BMDMs were pretreated with 10 mM 3-MA for 3 h and 20 μg ml−1 anti-MHC-II (I-A/I-E, M5/114 monoclonal antibody by Bio X Cell) or 20 μg ml−1 anti-IFNβ (BioLegend, 508107) for 24 h where necessary. Samples were imaged with a Leica M205 FA dissection microscope, and spots were counted using ImageJ. The peptides used for the assays were as follows: H1-1: KKPKVVKAKKVAKSPA, RPL30: PGDSDIIRSMPEQTGEK, COL1A: TPAKNSYSRAQANKH, HRNRPL: YGNVEKVKFMKSKPG, EXOSC4: GPHEIRGSRSRALPD.
Autophagosome profiling coupled to mass spectrometry
U2-OS cells constitutively expressing APEX2−Flag−LC3B were subjected to autophagosome content profiling as described in Le Gerroué et al.62. Quantification of the proteasomal content of autophagosomes via mass spectrometry was based on SILAC. In brief, APEX2−Flag−LC3B proximal proteins were biotinylated in ‘medium’ and ‘heavy’ labeled cells by inducing APEX2 activity after incubation with biotinphenol (500 mM) for 2 hat 37 °C and a 1-mimpulse with H2O2 (1 mM). Biotinphenol was also added to ‘light’ labeled cells, but biotinylation was not induced by omission of H2O2 application. In heavy labeled cells, autophagy was induced with genotoxic stress for 16 h of etoposide treatment (10 µM), whereas medium and light labeled cells were treated with DMSO. Autophagosome enrichment was induced by adding BafA (200 nM) to all cells for 2 h simultaneously to the biotinphenol treatment. Quenching solution (1 mM sodium azide, 10 mM sodium ascorbate and 5 mM Trolox in DPBS) was added to all cells to stop remaining biotinylation reactions, followed by three washing steps with PBS.
Cells were collected from 15-cm dishes by trypsinization (3 min, 37 °C). Afterwards, cells were washed twice in PBS prior to mixing them in a 1:1:1 ratio based on cell numbers.
An autophagosome-enriched fraction was recovered from the cells by the steps below performed at 4 °C prior to a streptavidin pulldown. Cells were washed and incubated in homogenization buffer I (10 mM KCl, 1.5 mM MgCl2, 10 mM HEPES-KOH and 1 mM DTT (pH 7.5)) for 20 min in an overhead shaker. Afterwards, cells were transferred into a dounce homogenizer and lysed with tight-fitting pestle B. The lysate was transferred into a new reaction vessel and diluted in homogenization buffer II (75 mM KCl, 22.5 mM MgCl2, 220 mM HEPES-KOH and 0.5 mM DTT (pH 7.5)). After centrifugation, the autophagosome-rich supernatant was treated with proteinase K (30 µg ml−1) and 1 mM CaCl2 for 30 min. Then, 5 mM PMSF was added to inactivate proteinase K. The fraction was cleared by centrifugation at 17,000g for 15 min, and the pellet was resuspended and incubated in RIPA buffer (50 mM Tris-HCl (pH 7.4), 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS and 150 mM NaCl) for 30 min. Afterwards, the lysate was cleared by centrifugation at 20,000g for 15 min. The supernatant was incubated overnight with pre-equilibrated NeutrAvidin beads. Beads were washed four times with RIPA buffer, and proteins were eluted by boiling in 3× sample buffer supplemented with 1 mM DTT for 20 min at 95 °C. Proteins were incubated with CAA (4.5 mM) in the dark and resolved on SDS-PAGE. In-gel digestion using trypsin followed prior to subjection of the peptides to LC−MS/MS analysis.
Differential protein analysis was performed using rule-based (frequency of identification, fold change) and statistical tests (t-test). Uniquely identified proteins were marked as those identified in all three biological repeats of the group and in none of the repeats of the comparing condition. Commonly identified but differentially abundant proteins between the two conditions were reported as those with greater than or equal to 1.75-fold change of average protein abundance between groups and t-test P < 0.05, for proteins identified in all three biological repeats in each of the comparing conditions. The rest of the proteins were considered non-significant. The t-test was performed on log2-transformed protein intensity values between the heavy and medium isotopically labeled conditions. Analysis was performed in Python programming language using common libraries (scipy, statmodels, numpy, pandas and matplotlib).
Data visualization
Plots were created by a free online platform for data visualization, SRplot (https://www.bioinformatics.com.cn/en), and GraphPad Prism 8.0.
Statistics and reproducibility
All statistical analyses were performed in GraphPad Prism 8.0. Error bars indicate s.e.m. among replicates. Asterisks indicate the significance set at P value: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P < 0.0001 (two-tailed Student’s t-test). No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications34. No data were excluded from the analyses. Data distribution was assumed to be normal, but this was not formally tested. The investigators were not blinded to allocation during experiments and outcome assessment. Samples were allocated to experimental groups according to genotypes or treatments. No method of randomization was used to assign samples to experimental groups.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE106 partner repository with dataset identifiers PXD058775 and PXD058936. All data needed to evaluate the conclusions in this paper are present in the paper and/or the supplementary materials. All data supporting the findings are also available from the corresponding author upon reasonable request.
References
Chatzinikolaou, G., Karakasilioti, I. & Garinis, G. A. DNA damage and innate immunity: links and trade-offs. Trends Immunol. 35, 429–435 (2014).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Kell, L. et al. The central role of DNA damage in immunosenescence. Front. Aging 4, 1202152 (2023).
Schumacher, B. et al. The central role of DNA damage in the ageing process. Nature 592, 695–703 (2021).
Yousefzadeh, M. J. et al. An aged immune system drives senescence and ageing of solid organs. Nature 594, 100–105 (2021).
Zhao, Y. et al. DNA damage and repair in age-related inflammation. Nat. Rev. Immunol. 23, 75–89 (2023).
Coppe, J. P. et al. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010).
Hartlova, A. et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015).
Arvanitaki, E. S. et al. Microglia-derived extracellular vesicles trigger age-related neurodegeneration upon DNA damage. Proc. Natl Acad. Sci. USA 121, e2317402121 (2024).
Chatzidoukaki, O. et al. R-loops trigger the release of cytoplasmic ssDNAs leading to chronic inflammation upon DNA damage. Sci. Adv. 7, eabj5769 (2021).
Karakasilioti, I. et al. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 18, 403–415 (2013).
Uchihara, Y. et al. DNA damage promotes HLA class I presentation by stimulating a pioneer round of translation-associated antigen production. Mol. Cell 82, 2557–2570 (2022).
Sheu, K. M. & Hoffmann, A. Functional hallmarks of healthy macrophage responses: their regulatory basis and disease relevance. Annu. Rev. Immunol. 40, 295–321 (2022).
Park, M. D. et al. Macrophages in health and disease. Cell 185, 4259–4279 (2022).
Takahashi, R. et al. Attenuated phagocytosis of secondary necrotic neutrophils by macrophages in aged and SMP30 knockout mice. Geriatr. Gerontol. Int. 16, 135–142 (2016).
Rawji, K. S. et al. Deficient surveillance and phagocytic activity of myeloid cells within demyelinated lesions in aging mice visualized by ex vivo live multiphoton imaging. J. Neurosci. 38, 1973–1988 (2018).
Barman, P. K. et al. Production of MHCII-expressing classical monocytes increases during aging in mice and humans. Aging Cell 21, e13701 (2022).
Cui, C. Y. et al. Skewed macrophage polarization in aging skeletal muscle. Aging Cell 18, e13032 (2019).
Franceschi, C. et al. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018).
Admon, A. The biogenesis of the immunopeptidome. Semin. Immunol. 67, 101766 (2023).
Prinz, J. C. Immunogenic self-peptides - the great unknowns in autoimmunity: identifying T-cell epitopes driving the autoimmune response in autoimmune diseases. Front. Immunol. 13, 1097871 (2022).
Kroemer, G., Marino, G. & Levine, B. Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010).
White, E., Lattime, E. C. & Guo, J. Y. Autophagy regulates stress responses, metabolism, and anticancer immunity. Trends Cancer 7, 778–789 (2021).
Munz, C. Autophagy beyond intracellular MHC class II antigen presentation. Trends Immunol. 37, 755–763 (2016).
Rock, K. L., Reits, E. & Neefjes, J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 37, 724–737 (2016).
Li, Y. et al. Nuclear accumulation of UBC9 contributes to SUMOylation of lamin A/C and nucleophagy in response to DNA damage. J. Exp. Clin. Cancer Res. 38, 67 (2019).
Mucino-Hernandez, G. et al. Nucleophagy contributes to genome stability through degradation of type II topoisomerases A and B and nucleolar components. J. Cell Sci. 136, jcs260563 (2023).
Kamileri, I., Karakasilioti, I. & Garinis, G. A. Nucleotide excision repair: new tricks with old bricks. Trends Genet. 28, 566–573 (2012).
Apostolou, Z. et al. Nucleotide excision repair and transcription-associated genome instability. Bioessays 41, e1800201 (2019).
Elango, R. et al. The structure-specific endonuclease complex SLX4−XPF regulates Tus−Ter-induced homologous recombination. Nat. Struct. Mol. Biol. 29, 801–812 (2022).
Kumar, N., Raja, S. & Van Houten, B. The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage. Nucleic Acids Res. 48, 11227–11243 (2020).
Zhu, X. D. et al. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12, 1489–1498 (2003).
Li, T. T. & Vasquez, K. M. Multi-faceted roles of ERCC1-XPF nuclease in processing non-B DNA structures. DNA 2, 231–247 (2022).
Goulielmaki, E. et al. Tissue-infiltrating macrophages mediate an exosome-based metabolic reprogramming upon DNA damage. Nat. Commun. 11, 42 (2020).
Nusser, A. et al. The development of autoimmune features in aging mice is closely associated with alterations of the peripheral CD4+ T-cell compartment. Eur. J. Immunol. 44, 2893–2902 (2014).
Baaten, B. J., Li, C. R. & Bradley, L. M. Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 3, 508–512 (2010).
Hengel, R. L. et al. Cutting edge: L-selectin (CD62L) expression distinguishes small resting memory CD4+ T cells that preferentially respond to recall antigen. J. Immunol. 170, 28–32 (2003).
Chatila, T. et al. Mechanisms of T cell activation by the calcium ionophore ionomycin. J. Immunol. 143, 1283–1289 (1989).
Oda, T. et al. DNA-double strand breaks enhance the expression of major histocompatibility complex class II through the ATM-NF-κB-IRF1-CIITA pathway. Cancer Gene Ther. 29, 225–240 (2022).
Lhuillier, C. et al. Radiotherapy-exposed CD8+ and CD4+ neoantigens enhance tumor control. J. Clin. Invest. 131, e138740 (2021).
Marin, I. et al. Cellular senescence is immunogenic and promotes antitumor immunity. Cancer Discov. 13, 410–431 (2023).
Ding, J. et al. Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol. Sci. 27, 338–344 (2006).
van Maanen, J. M. et al. Mechanism of action of antitumor drug etoposide: a review. J. Natl Cancer Inst. 80, 1526–1533 (1988).
Curtsinger, J. M., Lins, D. C. & Mescher, M. F. Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J. Exp. Med. 197, 1141–1151 (2003).
Curtsinger, J. M. & Mescher, M. F. Inflammatory cytokines as a third signal for T cell activation. Curr. Opin. Immunol. 22, 333–340 (2010).
Rahimi, R. A. & Luster, A. D. Chemokines: critical regulators of memory T cell development, maintenance, and function. Adv. Immunol. 138, 71–98 (2018).
Turley, S. J., Fletcher, A. L. & Elpek, K. G. The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nat. Rev. Immunol. 10, 813–825 (2010).
Koncz, B., Balogh, G. M. & Manczinger, M. A journey to your self: the vague definition of immune self and its practical implications. Proc. Natl Acad. Sci. USA 121, e2309674121 (2024).
Cibrian, D. & Sanchez-Madrid, F. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol. 47, 946–953 (2017).
Richert, I. et al. A TLR4 agonist induces osteosarcoma regression by inducing an antitumor immune response and reprogramming M2 macrophages to M1 macrophages. Cancers (Basel) 15, 4635 (2023).
Pisetsky, D. S. Pathogenesis of autoimmune disease. Nat. Rev. Nephrol. 19, 509–524 (2023).
Roche, P. A. & Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015).
Cardoso, M. H. et al. Impaired lysosome reformation in chloroquine-treated retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 64, 10 (2023).
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Murley, A. & Dillin, A. Macroautophagy in quiescent and senescent cells: a pathway to longevity? Trends Cell Biol. 33, 495–504 (2023).
Feng, Z. et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl Acad. Sci. USA 102, 8204–8209 (2005).
Valentin-Vega, Y. A. et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood 119, 1490–1500 (2012).
Bartsch, K. et al. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet. 26, 3960–3972 (2017).
Lan, Y. Y. et al. Extranuclear DNA accumulates in aged cells and contributes to senescence and inflammation. Aging Cell 18, e12901 (2019).
Hewitt, G. & Korolchuk, V. I. Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends Cell Biol. 27, 340–351 (2017).
Juretschke, T. & Beli, P. Causes and consequences of DNA damage-induced autophagy. Matrix Biol. 100-101, 39–53 (2021).
Le Guerroué, F. et al. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol. Cell 68, 786–796 (2017).
Mauthe, M. et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455 (2018).
Mauvezin, C. & Neufeld, T. P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11, 1437–1438 (2015).
Liang, Q. et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15, 228–238 (2014).
Zhao, M. et al. CGAS is a micronucleophagy receptor for the clearance of micronuclei. Autophagy 17, 3976–3991 (2021).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Macia, E. et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10, 839–850 (2006).
Tan, J. X. & Finkel, T. Lysosomes in senescence and aging. EMBO Rep. 24, e57265 (2023).
Rovira, M. et al. The lysosomal proteome of senescent cells contributes to the senescence secretome. Aging Cell 21, e13707 (2022).
Taieb, A. et al. Clinical, immunological, and genetic investigations in a rare association of type 1 diabetes with xeroderma pigmentosum. Pediatr. Endocrinol. Diabetes Metab. 28, 233–237 (2022).
Nance, M. A. & Berry, S. A. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 42, 68–84 (1992).
Ozdirim, E. et al. Cockayne syndrome: review of 25 cases. Pediatr. Neurol. 15, 312–316 (1996).
Zayoud, K. et al. Immunity in the progeroid model of Cockayne syndrome: biomarkers of pathological aging. Cells 13, 402 (2024).
Ocadlikova, D. et al. Chemotherapy-induced tumor cell death at the crossroads between immunogenicity and immunotolerance: focus on acute myeloid leukemia. Front. Oncol. 9, 1004 (2019).
Broecker, F. et al. Chemotherapy-induced, broadly reactive autoantibodies in a colon cancer patient. Cureus 14, e31954 (2022).
Perricone, C. et al. Smoke and autoimmunity: the fire behind the disease. Autoimmun. Rev. 15, 354–374 (2016).
Harel-Meir, M., Sherer, Y. & Shoenfeld, Y. Tobacco smoking and autoimmune rheumatic diseases. Nat. Clin. Pract. Rheumatol. 3, 707–715 (2007).
Meas, R., Burak, M. J. & Sweasy, J. B. DNA repair and systemic lupus erythematosus. DNA Repair (Amst.) 56, 174–182 (2017).
Micheli, C. et al. UCTD and SLE patients show increased levels of oxidative and DNA damage together with an altered kinetics of DSB repair. Mutagenesis 36, 429–436 (2021).
Stergioti, E. M. et al. Transcriptomic and proteomic profiling reveals distinct pathogenic features of peripheral non-classical monocytes in systemic lupus erythematosus. Clin. Immunol. 255, 109765 (2023).
Manolakou, T. et al. ATR-mediated DNA damage responses underlie aberrant B cell activity in systemic lupus erythematosus. Sci. Adv. 8, eabo5840 (2022).
Westhorpe, C. L. V. et al. Effector CD4+ T cells recognize intravascular antigen presented by patrolling monocytes. Nat. Commun. 9, 747 (2018).
Liu, F. et al. Distinct fate, dynamics and niches of renal macrophages of bone marrow or embryonic origins. Nat. Commun. 11, 2280 (2020).
Siametis, A. et al. Transcription stress at telomeres leads to cytosolic DNA release and paracrine senescence. Nat. Commun. 15, 4061 (2024).
Deretic, V. Autophagy in inflammation, infection, and immunometabolism. Immunity 54, 437–453 (2021).
Dengjel, J. et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl Acad. Sci. USA 102, 7922–7927 (2005).
Neefjes, J. et al. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 (2011).
Alissafi, T. et al. Tregs restrain dendritic cell autophagy to ameliorate autoimmunity. J. Clin. Invest. 127, 2789–2804 (2017).
Keller, C. W. et al. ATG-dependent phagocytosis in dendritic cells drives myelin-specific CD4+ T cell pathogenicity during CNS inflammation. Proc. Natl Acad. Sci. USA 114, E11228–E11237 (2017).
Romao, S. et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J. Cell Biol. 203, 757–766 (2013).
Ma, J. et al. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J. Biol. Chem. 287, 34149–34156 (2012).
Lee, H. K. et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239 (2010).
Clement, C. C. et al. Pleiotropic consequences of metabolic stress for the major histocompatibility complex class II molecule antigen processing and presentation machinery. Immunity 54, 721–736 (2021).
Fugmann, T. et al. The MHC class II immunopeptidome of lymph nodes in health and in chemically induced colitis. J. Immunol. 198, 1357–1364 (2017).
Schrezenmeier, E. & Dorner, T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16, 155–166 (2020).
Li, W. et al. Low-dose chloroquine treatment extends the lifespan of aged rats. Protein Cell 13, 454–461 (2022).
Doeppner, T. R. et al. Long-term treatment with chloroquine increases lifespan in middle-aged male mice possibly via autophagy modulation, proteasome inhibition and glycogen metabolism. Aging (Albany NY) 14, 4195–4210 (2022).
Qian, M. et al. Boosting ATM activity alleviates aging and extends lifespan in a mouse model of progeria. eLife 7, e34836 (2018).
Liu, A. et al. Autophagy inhibitor 3-methyladenine mitigates the severity of colitis in aged mice by inhibiting autophagy. Exp. Gerontol. 201, 112697 (2025).
Bujarrabal-Dueso, A. et al. The DREAM complex functions as conserved master regulator of somatic DNA-repair capacities. Nat. Struct. Mol. Biol. 30, 475–488 (2023).
Yu, X. et al. Therapeutic potential of tolerance-based peptide vaccines in autoimmune diseases. Int. Immunopharmacol. 116, 109740 (2023).
Marino, F. et al. High-throughput, fast, and sensitive immunopeptidomics sample processing for mass spectrometry. Methods Mol. Biol. 1913, 67–79 (2019).
Zolg, D. P. et al. INFERYS rescoring: boosting peptide identifications and scoring confidence of database search results. Rapid Commun. Mass Spectrom. 39, e9128 (2021).
Kall, L. et al. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 (2007).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Acknowledgements
The Horizon 2020 Marie Curie ITN ‘HealthAge’ (GA 812830); the Horizon-WIDERA-2023-Talents-01-01: ERA Chairs ‘RACE’ (1011803090); the Horizon-WIDERA-Access Pathways to Synergies 2023 ‘TRIAD’ (101158508); the Horizon-WIDERA-2023-Talents-01, ERA Chair InflaCare (101180309); the Fondation Santé; ELIDEK grant 11578; ELIDEK PhD fellowships 11034 and 11330; the Hevolution Foundation; the Uni-Pharma Kleon Tsetis Pharmaceutical Laboratories S.A. (PAR00838) and Pharmathen S.A. (PAR00863) research funds; and the Greece 2.0, National Recovery and Resilience Plan Flagship program TAEDR-0535850 supported this work. The research in the Beli laboratory was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation; Project-ID 259130777 – SFB 1177).
Author information
Authors and Affiliations
Contributions
Conceptualization: G.N., E.A. and G.A.G. Methodology and investigation: G.N., E.A., E.T., A.S., T.J., O.A., K.T., G.B., E.D. and P.B. Visualization: G.N. and E.A. Supervision: G.A.G. Writing—original draft: G.N., E.A. and G.A.G. Writing—review and editing: G.N., E.A. and G.A.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Immune complex deposition and glomerular damage in the kidneys of mice with an Ercc1 defect in monocyte-derived macrophages.
Immunofluorescence analysis of kidney cryosections stained with antibodies raised against IgM and IgA immunoglobulins or C3 complement protein. Kidney glomeruli are shown in all images. The plots on the bottom depict the mean fluorescence intensity (MFI) of each staining (n = 5-7, pval = 0.0039 and pval = 0.0327). (B.) Histological analysis (H&E staining) of skins derived from 8-month-old wt or Er1Lyz2/− mice. Magnifications are indicated. (n = 4) Scale bars: 10μm. (C.) Caliper measurements of the thickness of paws and ankles from 8-month-old wt and Er1Lyz2/− animals (n = 6, pval > 0.05). (D.) PAS staining of kidney paraffin sections. The white arrow indicates glomerular damage (n = 3). (E.) ELISA for the detection of albumin in the urine of wt and Er1Lyz2/− animals (n = 5, pval > 0.05). (F.) Autoantibody detection in the sera of 8-month-old wt and Er1Lyz2/− mice using an indirect immunofluorescence assay. Primary wild-type mouse embryonic fibroblasts (MEFs) were seeded and incubated with the mouse sera, as indicated. Here, representative images of 1:50 sera dilutions are presented. The autoantibody positivity for mice is also expressed as the percentage indicated on the top right of each image (n = 10 biological replicates and 5 independent optical fields/mouse serum were assessed).(G.) Patterns of fluorescence in MEFs incubated with 8-month-old Er1Lyz2/− sera are indicated(n = 10). Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 2 Serum transfer experiement and gating strategy of immune cell populations examined in wt and Er1Lyz2/− animals.
(A.) Adoptive transfer of sera derived from wt or Er1Lyz2/− animals to young wt hosts. A schematic diagram is shown (top). The immunofluorescence images depict glomeruli from kidney cryosections, stained with an IgM antibody (bottom) (n = 6, pval = 0.0143). (B.) H&E analysis of spleens isolated from 8-month-old wt and Er1Lyz2/− animals. The magnification is indicated (n = 6-7). (C.) Weights of spleens isolated from 8-month-old wt and Er1Lyz2/− animals (n = 8, pval = 0.0174). (D.) Flow cytometry analysis of live splenocytes. Gating sequence for analysis of plasma cells in wt and Er1Lyz2/− splenocytes. The plot shows the percentages of CD19+CD138+ plasma cells (n = 3, pval = 0.0293). (E.) Bar charts of the number of CD19+CD138+ plasma cells in the lymph nodes and bone marrows of 8-month-old mice (n = 4-5, pval = 0.0295). (F-I.) Gating sequence for analysis of monocyte-derived cells (F.), CD4+ T cells (G.) cytotoxic NK and CD8+ cells (H.) and regulatory CD4+ T cells (I.). Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 3 Activated monocytic origin cells and CD4+ T cells in Er1Lyz2/− and in naturally aged wt mice.
(A.) Flow cytometry analysis of bone marrows derived from 8-month-old wt and Er1Lyz2/− animals for the detection of myeloid granulocyte-monocyte progenitors. The scatter plots show the gating sequence and representative images. The bar plot shows the quantification of the Lin−c-kit+Sca-1−CD34+CD16/32+ cell population (n = 5). (B.) Gating sequence for the flow cytometry analysis of splenic macrophages and representative plots for the quantification of the MHC-II MFIs (n = 3, pval = 0.0474). (C.) MHC-II (left) and CD86 (right) protein expression levels in wt and Er1Lyz2/− splenic macrophages. The overlaying histograms represent the fluorescence intensities of wt, Er1Lyz2/− or isotype (iso) control samples (in the dashed black line, continuous black line and in gray color respectively). Mean fluorescence intensities are plotted in the bar graphs (n = 4-6, pval = 0.0283 and pval = 0.0261). (D.) Numbers of memory CD4+ T cells (CD44hiCD62Ll°) in wt and Er1Lyz2/− spleens (n = 5, pval = 0.026). (E.) Representative scatter plots and corresponding bar charts depicting the percentage of activated CD4+ T cells (CD44hiCD62Ll°) in 8- and 10-month-old wt and Er1Lyz2/− inguinal lymph nodes (n = 3, pval=0.031 and pval=0.0276). (F.) Plots showing the percentages of naïve CD4+ T cells (CD44l°CD62Lhi) in 2- and 10-month-old spleens (n = 4-6, pval = 0.0131). (G.) Representative scatter plots of CD44 and CD62L expression in 2- (young) and 24-month-old (aged) wt spleens (n = 4). (H.) Representative histogram plots of T-bet protein levels in CD4+ T cells after staining for T-bet and CD4 in 10-month-old wt and Er1Lyz2/− lymph node single cell suspensions (shown in the dashed black line and continuous black line respectively). (I.) Histograms overlaying the expression of ki67 protein in CD4+CD44+ cells in splenocytes isolated from 10-month-old wt and Er1Lyz2/− animals and stained for the ki67 proliferation, CD4 T cell and CD44 activation marker (shown in gray and in the continuous black line respectively) (n = 4, pval = 0.0487). Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05 (two-tailed Student’s t-test).
Extended Data Fig. 4 Increased kidney infiltration and adaptive immune cell activation in T-cell and BMDM transfer experiments respectively.
(A.) Schematic diagram showing the CD4+ T-cell transfer experiment. (B.) H&E staining of kidney paraffin sections derived from wt NSG hosts receiving wt or and Er1Lyz2/− CD4+ T cells. T cells were isolated from 8-10-month-old animals. The arrows show the focal inflammation observed in all the kidneys of mice receiving Er1Lyz2/− CD4+ T cells (n = 6). Representative images from all individual mice (left) and a higher magnification image (right, 400x magnification) are depicted. (C.) Gating sequence for the analysis of CD4+ T cells in kidney Percoll fractions. The bar plot shows the quantification of CD4+ T cell numbers localized in the kidneys of animals receiving wt or Er1Lyz2/− T cells (n = 5, pval = 0.0477). (D-E.) Representative plots of (D.) activated CD4+T cells and (E.) plasma cells in the spleens of animals receiving wt or Er1Lyz2/− BMDMs. Plasma cell percentages are quantified (n = 4, pval = 0.0433). Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05 (two-tailed Student’s t-test). Scale bars: 100μm.
Extended Data Fig. 5 MHC-II dependence on the DDR in Er1Lyz2/− BMDMs and in NZB/W monocytes.
(A.) Immunofluorescence analysis for the detection of DNA damage levels in wt untreated, Er1Lyz2/− untreated and Er1Lyz2/− anti-oxidant-treated (mito-TEMPO and N-acetylcysteine (NAC)) BMDMs. The percentage of cells with 3 or more γ-H2A.X+53BP1+ co-localized foci is plotted (n = 3-4, pval = 0.0036, pval = 0.0204 and pval = 0.0093). Single-channel images of Extended Data Fig. 5A are shown in Supplementary file 2D(B.) Immunofluorescence analysis for the detection of 8-oxoguanine (8-oxoG) lesions in wt and Er1Lyz2/− BMDMs. The plot shows the MFI of 8-oxoG in each condition (n = 4, pval = 0.0086). (C.) Western blot analysis for the quantification of phosphorylated ATM and total ATM levels in whole-cell extracts. Beta tubulin was used for normalization (n = 4-5, pval = 0.0307). (D.) Gating strategy for the flow cytometry analysis of single cell suspensions of BMDMs and MHC-II levels in wt and Er1Lyz2/− untreated macrophages and Er1Lyz2/− cells treated with an ATR or a DNA-PK inhibitor (n = 3). (E.) Flow cytometry analysis of the quantification of γ-H2A.X levels in cells of monocytic origin isolated from lupus prone NZB/W 11-month-old mice (SLE) or from their age-matched controls (cont). An isotype control histogram is shown in gray (n = 4, pval = 0.0365). (F.) Quantification of the MHC-II levels in control, lupus prone untreated (SLE) and lupus monocyte-derived cells treated with an ATM kinase inhibitor (SLE+ATMi). The MFI in each corresponding condition is plotted (n = 5-6, pval = 0.0037 and pval = 0.0074). (G.) Histograms showing the expression of MHC-II protein in wt BMDMs treated with camptothecin (wtCPT, left), tunicamycin or underwent nutrient starvation (wttun and wtstarv respectively, right), as indicated, and their corresponding plots for their quantification (n = 3-4, pval = 0.0138). Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 6 Type I IFN signaling in Er1Lyz2/− BMDMs and in vivo antibody blockade experiments.
(A.) Immunofluorescence detection of the colocalized DNA damage markers γ-H2A.X and 53BP1 in wt untreated and wt etoposide-treated BMDMs. The percentage of positive cells for over 2 colocalized γ-H2A.X+53BP1+ foci/cell is plotted. (At least 4 independent optical fields were counted from n = 3 biological replicates, pval = 0.0018) Single-channel images of Extended Data Fig. 6A are shown in Supplementary file 3A(B.) Quantitation of the mRNA levels of chemokine genes in Er1Lyz2/− BMDMs, as shown. The mRNA levels of these genes in wt BMDMs are indicated with the red dotted line (n = 3-6, exact pvalue provided in Source Data file). (C.) (Top) Western blotting for the detection of interferon beta (IFN-β) protein levels secreted in the supernatants of wt and Er1Lyz2/− BMDMs. The splice point is indicated by a vertical black line. Equal volumes of concentrated supernatants were loaded on the gel. For the normalization of the secreted protein levels, equal volumes of cells lysed with RIPA buffer were loaded and the membranes were probed with beta tubulin. (Bottom) Blots for the quantification of interferon regulatory factor 5 (IRF5) and phosphorylated signal transducer and activator of transcription 1 (pSTAT1) versus total STAT1 protein levels. Actin was used for normalization (n = 3-4, pval = 0.0011 for IFN-β, pval= 0.009 for STAT1, pval = 0.048948 for IRF5). (D.) Western blot analysis of ERCC1 protein levels in CD4+ T cells purified from wt or Er1Lyz2/− mice at the age of 10 months (n = 3). (E.) Gating strategy for the flow cytometry analysis of CD4+ T cells in a BMDM-CD4+ T-cell co-culture. (F-G.) Flow cytometry analysis of splenocytes isolated from wt and Er1Lyz2/− anti-MHC-II-treated mice and from wt and Er1Lyz2/− isotype control-treated mice and stained for (F.) the CD44 and CD62L activation T-cell markers and (G.) the CD11b myeloid cell marker and MHC-II. MHC-II levels exhibit a drop in the CD11b+ population (n = 3, pval = 0,0384). (H.) Immunofluorescence analysis of C3 complement protein localized in the kidney glomeruli of Er1Lyz2/− isotype control-treated and Er1Lyz2/− anti-MHC-II-treated animals. Representative images and plots with the MFI quantification of C3 are shown (n = 3, pval = 0.0343). (I.) H&E staining of kidneys derived from Er1Lyz2/− anti-CD4-and isotype control-treated animals. The arrows point to inflammatory foci (n = 6-8, pval = 0,0452). Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01, ***≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 7 The antigenic landscape of wt BMDMs treated with LPS.
(A.) Immunofluorescence staining of γ-Η2A.X and 53BP1 in wt untreated, wt LPS-treated (wtLPS) and Er1Lyz2/− BMDMs. Cells with over 2 colocalized foci of the two proteins were labeled positive and indicated using white arrows. The percentage of positive cells is plotted. (At least 4 independent optical fields were counted from n = 3 biological replicates.) Single-channel images of Extended Data Fig. 7A are shown in Supplementary file 3B (B.) Flow cytometry analysis of the MHC-II expression levels in wt untreated, wtLPS and Er1Lyz2/− BMDMs. Representative histograms and MFIs are plotted (n = 3-5). (C.) Immunoprecipitation and western blot detection of the MHC-II protein in BMDMs using an anti-MHC-II (IP) or isotype control antibody (IgG). (D.) Volcano plot of differentially presented peptides in wt (downregulated, blue) and wtLPS cells (upregulated, red). Log2(Fold Change) of -6 or 6 represents peptides uniquely identified in the wt or wtLPS MHC-peptidome. Statistical significance (ANOVA analysis) was set at p-value ≤ 0.05 (horizontal black dashed line) and peptide enrichment at Log2(Fold Change)≥0.3 (upregulated in wtLPS) or Log2(Fold Change)≤-0.3 (downregulated in wtLPS) (vertical black dashed line). Extracellular matrix proteins are labeled with their corresponding gene symbol. (E.) Bubble plot of the Gene Ontology (GO) term enrichment analysis (cellular component, Mann-Whitney U test) of significantly over-represented wtLPS peptides, when compared to wt controls (p-value ≤ 0.05 and Log2Fold change≥0.3). The dot size shows the total count of genes per annotated pathway and the blue color scale indicates the statistical significance as per the p-value of the enriched pathways. The x axis indicates the fold enrichment derived from pathway analysis. (F.) IFN-γ ELISpot analysis of splenocytes isolated from either 8-month-old wt or Er1Lyz2/− mice and pulsed with the indicated peptides. Representative images are shown. Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 8 ATR-dependent Lamin B1 loss and increased autophagic flux in Er1Lyz2/− BMDMs.
(A.) Cell cycle analysis of wt and Er1Lyz2/− BMDMs stained with DAPI-ki67. The rectangle gates in the representative scatter plots and the bar chart shows the percentages of cells in the indicated cell cycle phases (n = 4). (B.) Western blotting of whole-cell extracts derived from wt and Er1Lyz2/− untreated cells and cells treated with ATM and ATR inhibitors for the quantification of Lamin B1. Actin was used as a loading control (n = 4, pval=0.0487 and pval= 0.0025). (C.) Immunofluorescence staining of γ-Η2AX and 53BP1 in Er1Lyz2/− untreated, 3-MA treated and CQ-treated BMDMs. Cells with over 2 colocalized foci of the two proteins were considered positive and the percentage of positive cells is plotted. (At least 5 independent optical fields were counted from n = 3 biological replicates.) Single-channel images of Extended Data Fig. 8C are shown in Supplementary file 3C. (D.) Immunofluorescence detection of cGAS in wt untreated, Er1Lyz2/− untreated and Er1Lyz2/− BMDMs that underwent FBS-starvation (Er1Lyz2/− +starv). The bar plot shows the MFI of cytoplasmic cGAS in the corresponding conditions. (At least 4 independent optical fields were counted from n = 3 biological replicates, pval=0.0095 and pval=0.0069). Single-channel images of Extended Data Fig. 8D are shown in Supplementary file 3B(E.) Autophagic flux measurement after lentiviral infection of wt and Er1Lyz2/− BMDMs with a construct for the expression of LC3-GFP-mCherry. Green fluorescence corresponds to autophagosomes, while red fluorescence represents the autophagosomes and lysosomes in each cell. The graph shows the ratio of average red to green fluorescence per cell. (At least 3 independent optical fields were counted from n = 4 biological replicates, pval=0.0170) (F-G.) An overlay of the fluorescence intensity of (F.) LysoTracker Red (n = 5, pval=0.0289) and (G.) LysoSensor Green (n = 7-8, pval=0.0302) in Er1Lyz2/− BMDMs and wt corresponding controls, measured by flow cytometry analysis. The graphs show the respective MFI values. Error bars indicate S.E.M. among replicates. Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 9 Lack of cell death in Er1Lyz2/− BMDMs and HLA-DR levels in THP-1 cells.
(A.) Immunofluorescence staining of early endosome antigen 1 (EEA1) in Er1Lyz2/− untreated and dynasore-treated BMDMs. White insets indicate a higher magnification on the right (n = 3). (B.) Single- or two-channel images and higher magnifications of Fig. 5a. Magenta arrowheads point to H1+p62+LAMP-1+ foci. White arrows point to cytoplasmic chromatin fragments. (C.) Flow cytometry analysis of wt and Er1Lyz2/− BMDMs stained with annexin V and propidium iodide (PI) for the detection of cell death. Scatter plots are and the percentages of annexin−PI− cells are shown (n = 4). (D.) LAMP-1 protein levels in equally loaded lysosomal extracts isolated from wt and Er1Lyz2/− BMDMs. Ponceau stain was used for normalization. (n = 4, pval=0.0147) (E.) Quantification of MHC-II in untreated and etoposide-treated THP-1 human monocyte cells. An isotype control histogram is shown in gray (n = 4, pval=0.0006). Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01, ***≤0.01 (two-tailed Student’s t-test). Scale bars: 10μm.
Extended Data Fig. 10 DKO immunophenotyping and cGAS levels.
(A.) Western blotting for the detection of ERCC1 and ATG5 protein levels in wt and DKO (Ercc1fl/−; Atg5fl/fl; Lyz2-Cre+) BMDMs. The bar charts indicate the protein levels in the DKO cells while the red dotted line marks wt protein levels for comparison. GAPDH or actin were used for normalization (n = 4 pval<0,0001 and pval<0,0001). The splice point is indicated by a vertical black line (B.) H&E analysis of DKO spleens (n = 4). (C.) Quantification of macrophage numbers in the spleens of 8-month-old wt, Er1Lyz2/− and DKO mice (n = 4, pval = 0,0334). (D.) Detection of granulocyte-monocyte progenitors in the bone marrows of 8-month-old wt, Er1Lyz2/− and DKO mice. The red dotted line in the graph marks the mean percentage of GMPs in Er1Lyz2/− mice, comparable to the one in wt mice (the individual points of the measurements of wt and Er1Lyz2/− GMPs are shown in Supplementary Figure S3A) (n = 6). (E.) Representative scatter plots of the plasma cells in splenocytes isolated from 8-month-old wt, Er1Lyz2/− and DKO mice. The rectangle gate shows the percentage of CD19+CD138+ plasma cells. Total numbers are shown in Fig. 6d. (F.) Flow cytometry analysis of wt and DKO BMDMs stained for annexin V and PI. The percentage of live (annexin−PI−) cells is plotted (n = 3). (G.) Immunofluorescence staining of cGAS in wt, Er1Lyz2/− and DKO BMDMs. The MFIs are plotted (n = 4-6, pval = 0,0010 and pval = 0,0140). (H.) IFN-γ ELISpot of 1:4 BMDM: CD4+ T cell co-cultures: Er1Lyz2/− or DKO BMDMs were mixed with Er1Lyz2/− CD4+ T cells. The numbers of IFN-γ spots are plotted (n = 4, pval = 0,0091). Asterisk indicates the significance set at p-value: *≤0.05, **≤0.01, ****<0.0001 (two-tailed Student’s t-test). Scale bars: 10μm.
Supplementary information
Supplementary Information
Supplementary Figs. 1−4 and legends for Supplementary Figs. 1−4
Supplementary Table 1
Peptides identified in immunopeptidomics analysis. The peptides that were classified as low or high binders of MHC-II from all samples were included for this analysis (NetMHCIIpan version 4.3, H-2-IAb and H-2-IAq alleles).
Supplementary Table 2
Proteins identified in the autophagosomes of DMSO-treated or etoposide-treated U2-OS cells
Supplementary Table 3
Peptides identified in immunopeptidomics analysis of DKO and WT control BMDMs. The peptides that were classified as low or high binders of MHC-II from all samples were included for this analysis (NetMHCIIpan version 4.3, H-2-IAb and H-2-IAq alleles). The peptides with fold change > 1.5, P < 0.05 and group profile ratio >0.85:1.15 or >1.15:0.85 were taken into consideration.
Supplementary Data 1
Unprocessed western blots and/or gels for Supplementary Fig. 1
Supplementary Data 1
Statistical Source Data for Supplementary Fig. 1
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 2
Unprocessed western blots and/or gels
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 4
Unprocessed western blots and/or gels
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 5
Unprocessed western blots and/or gels
Source Data Fig. 6
Statistical Source Data
Source Data Fig. 6
Unprocessed western blots and/or gels
Source Data Fig. 7
Statistical Source Data
Source Data Fig. 7
Unprocessed western blots and/or gels
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 5
Unprocessed western blots and/or gels
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 6
Unprocessed western blots and/or gels
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 7
Unprocessed western blots and/or gels
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 8
Unprocessed western blots and/or gels
Source Data Extended Data Fig. 9
Statistical Source Data
Source Data Extended Data Fig. 9
Unprocessed western blots and/or gels
Source Data Extended Data Fig. 10
Statistical Source Data
Source Data Extended Data Fig. 10
Unprocessed western blots and/or gels
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Niotis, G., Arvanitaki, E.S., Theodorakis, E. et al. DNA damage in macrophages drives immune autoreactivity via nuclear antigen presentation. Nat Aging (2026). https://doi.org/10.1038/s43587-025-01053-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43587-025-01053-3