Abstract
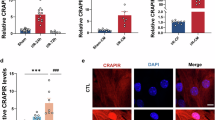
Targeting the cardiomyocyte cell cycle is a promising strategy for heart repair following injury. Here, we identify a cardiac-regeneration-associated PIWI-interacting RNA (CRAPIR) as a regulator of cardiomyocyte proliferation. Genetic ablation or antagomir-mediated knockdown of CRAPIR in mice impairs cardiomyocyte proliferation and reduces heart regenerative potential. Conversely, overexpression of CRAPIR promotes cardiomyocyte proliferation, reduces infarct size and improves heart function after myocardial infarction. Mechanistically, CRAPIR promotes cardiomyocyte proliferation by competing with NF110 for binding to the RNA-binding protein PA2G4, thereby preventing the interaction of PA2G4 with the NF110–NF45 heterodimer and reducing NF110 degradation. The ability of CRAPIR to promote proliferation was confirmed in human embryonic stem cell-derived cardiomyocytes. Notably, CRAPIR serum levels are lower in individuals with ischemic heart disease and negatively correlate with levels of N-terminal pro-brain natriuretic peptide. These findings position CRAPIR both as a potential diagnostic marker for cardiac injury and as a therapeutic target for heart regeneration through the PA2G4–NF110–NF45 signaling axis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA sequencing raw data generated in this study have been deposited in Sequence Read Archive at the NCBI Center under accession number PRJNA1186848. Other data are available in the main article and Supplementary Information. Source data are provided with this paper.
References
Poss, K. D., Wilson, L. G. & Keating, M. T. Heart regeneration in zebrafish. Science 298, 2188–2190 (2002).
Singh, B. N. et al. A conserved HH-Gli1-Mycn network regulates heart regeneration from newt to human. Nat. Commun. 9, 4237 (2018).
Senyo, S. E. et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436 (2013).
Bergmann, O. et al. Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 (2009).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011).
Nakada, Y. et al. Hypoxia induces heart regeneration in adult mice. Nature 541, 222–227 (2017).
Han, C. et al. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res. 25, 1137–1151 (2015).
Notari, M. et al. The local microenvironment limits the regenerative potential of the mouse neonatal heart. Sci. Adv. 4, eaao5553 (2018).
Ye, L. et al. Early regenerative capacity in the porcine heart. Circulation 138, 2798–2808 (2018).
Yokota, T. et al. Type V collagen in scar tissue regulates the size of scar after heart injury. Cell 182, 545–562 (2020).
Mohamed, T. M. A. et al. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 173, 104–116 (2018).
Cheng, Y. Y. et al. Metabolic changes associated with cardiomyocyte dedifferentiation enable adult mammalian cardiac regeneration. Circulation 146, 1950–1967 (2022).
Cho, K. W. et al. Polycomb group protein CBX7 represses cardiomyocyte proliferation through modulation of the TARDBP/RBM38 axis. Circulation 147, 1823–1842 (2023).
Feng, J. et al. Versican promotes cardiomyocyte proliferation and cardiac repair. Circulation 149, 1004–1015 (2024).
Gong, R. et al. Cyclin L1 controls cardiomyocyte proliferation and heart repair after injury. Signal Transduct. Target. Ther. 8, 243 (2023).
Wang, L. L. et al. Local and sustained miRNA delivery from an injectable hydrogel promotes cardiomyocyte proliferation and functional regeneration after ischemic injury. Nat. Biomed. Eng. 1, 983–992 (2017).
Chen, J. et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 112, 1557–1566 (2013).
Raso, A. et al. A microRNA program regulates the balance between cardiomyocyte hyperplasia and hypertrophy and stimulates cardiac regeneration. Nat. Commun. 12, 4808 (2021).
Borden, A. et al. Transient introduction of miR-294 in the heart promotes cardiomyocyte cell cycle reentry after injury. Circ. Res. 125, 14–25 (2019).
Ponnusamy, M. et al. Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation 139, 2668–2684 (2019).
Cai, B. et al. The long noncoding RNA CAREL controls cardiac regeneration. J. Am. Coll. Cardiol. 72, 534–550 (2018).
Cai, B. et al. Targeting LncDACH1 promotes cardiac repair and regeneration after myocardium infarction. Cell Death Differ. 27, 2158–2175 (2020).
Huang, S. et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139, 2857–2876 (2019).
Garikipati, V. N. S. et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 10, 4317 (2019).
Yan, J. et al. CircMap4k2 reactivated by aneurysm plication alleviates residual cardiac remodeling After SVR by enhancing cardiomyocyte proliferation in post-MI mice. J. Adv. Res. https://doi.org/10.1016/j.jare.2023.11.034 (2023).
Ma, W. et al. Oxidant stress-sensitive circRNA Mdc1 controls cardiomyocyte chromosome stability and cell cycle re-entry during heart regeneration. Pharmacol. Res. 184, 106422 (2022).
Ross, R. J., Weiner, M. M. & Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359 (2014).
Czech, B. et al. piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52, 131–157 (2018).
Han, H. et al. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 137, 1603–1614 (2021).
Ai, L. et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer 18, 88 (2019).
Gao, X. Q. et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nat. Cell Biol. 22, 1319–1331 (2020).
Wang, K. et al. HNEAP regulates necroptosis of cardiomyocytes by suppressing the m5C methylation of Atf7 mRNA. Adv. Sci. 10, e2304329 (2023).
Wang, K. et al. PIWI-interacting RNA HAAPIR regulates cardiomyocyte death after myocardial infarction by promoting NAT10-mediated ac4C acetylation of Tfec mRNA. Adv. Sci. 9, e2106058 (2022).
Ozata, D. M. et al. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 4, 156–168 (2020).
Dong, Z. W. et al. RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157 (2012).
Kim, C. K. et al. Negative regulation of p53 by the long isoform of ErbB3 binding protein Ebp1 in brain tumors. Cancer Res. 70, 9730–9741 (2010).
Ko, H. R. et al. P42 Ebp1 functions as a tumor suppressor in non-small cell lung cancer. BMB Rep. 48, 159–165 (2015).
Liu, Z., Ahn, J. Y., Liu, X. & Ye, K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc. Natl Acad. Sci. USA 103, 10917–10922 (2006).
Shen, Y. J. et al. Progression signature underlies clonal evolution and dissemination of multiple myeloma. Blood 137, 2360–2372 (2021).
Ye, J., Jin, H., Pankov, A., Song, J. S. & Blelloch, R. NF45 and NF90/NF110 coordinately regulate ESC pluripotency and differentiation. RNA 23, 1270–1284 (2017).
Haque, N., Will, A., Cook, A. G. & Hogg, J. R. A network of DZF proteins controls alternative splicing regulation and fidelity. Nucleic Acids Res. 51, 6411–6429 (2023).
Guan, D. et al. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell. Biol. 28, 4629–4641 (2008).
Yang, J., Xue, F. T., Li, Y. Y., Liu, W. & Zhang, S. Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur. Rev. Med. Pharmacol. Sci. 22, 7952–7961 (2018).
Huang, X. et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol. Cancer 18, 166 (2019).
Pilipenko, E. V. et al. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14, 2028–2045 (2000).
Wild, K. et al. MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments. Nat. Commun. 11, 776 (2020).
Hwang, I. et al. PA2G4/EBP1 ubiquitination by PRKN/PARKIN promotes mitophagy protecting neuron death in cerebral ischemia. Autophagy https://doi.org/10.1080/15548627.2023.2259215 (2023).
Nguyen, D. Q. et al. The role of ErbB3 binding protein 1 in cancer: friend or foe? J. Cell. Physiol. 233, 9110–9120 (2018).
Li, K. et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 30, 163–178 (2020).
Han, Z. et al. ALKBH5-mediated m6A mRNA methylation governs human embryonic stem cell cardiac commitment. Mol. Ther. Nucleic Acids 26, 22–33 (2021).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (92168119 to B.C., 82100300 and 82470266 to W.M., 82272389 to Yu Liu, 82300302 to Yining Liu, 82330011 and U21A20339 to B.Y.), the HMU Marshal Initiative Funding (HMUMIF-21018 to B.C.) and the key project of Natural Science Foundation of Heilongjiang Province (ZD2023H001 to N.W.).
Author information
Authors and Affiliations
Contributions
B.C. and W.M. conceived the study concept. H.C., W.H., Yanan Tian, Z.R., Xin Wang, J.L., Q.O., Y.H., H.J., X.L., XiuXiu Wang and Yining Liu performed the experimental studies. H.C., Yanan Tian and W.H. carried out the data analysis. B.C., W.M., Yu Liu, Ye Tian, F.L., N.W. and B.Y. wrote the manuscript. B.C., W.M. and N.W. responded to the editors’ and reviewers’ comments. B.C., W.M., Yu Liu, Yining Liu and B.Y. provided the funding. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended Data
Extended Data Fig. 1 The conservation and abundance of piRNAs.
a, The distribution of piRNA length. b, Heatmap of differentially expressed piRNAs in heart tissues. c, The blast of piRNA sequences between the mouse and human using NCBI’s blast tool. d, The expression of small RNAs in heart and cardiomyocytes (n=3, 4 biological replicates). The high expression of these three piRNAs was confirmed by comparing their expression levels to those of the well-known highly expressed miR-1 and miR-208 in the heart, and the rarely expressed miR-122 and miR-124, which are specifically expressed in liver or brain tissue, respectively. e, The expression level of AB352916, DQ540981 and AB351100 in postnatal day 1 (P1) or day 7 (P7) mouse heart tissues (n=3, 4 mice). Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with or without Welch’s correction (e). AR, apex resection.
Extended Data Fig. 2 The verification of piRNA AB352916 expression and features.
a, The expression of AB352916, DQ540981 and AB351100 in cardiomyocytes after transfecting with their antagomirs (n=4 independent experiment). b, The effects of DQ540981 and AB351100 antangomir on cardiomyocyte proliferation (n=3 independent experiment). c, The expression of AB352916 in cardiomyocytes after transfected with agomir (n=4 independent experiment). d, Detection of 2′‐O‐methylation at the 3′ end of AB352916 and miRNAs using RTL‐P approach. This experiment was repeated three times independently. e, The detection of the binding of PIWI proteins to CRAPIR using RNA pull down analysis and Western blot. This experiment was repeated three times independently. f, The expression of AB352916 in cardiomyocytes isolated from failing mouse hearts (n=5 biological replicates). g, The expression of AB352916 in different tissues (n=3 mice). h, The distribution of AB352916 between the nucleus and cytoplasm in cardiomyocytes (n=3 biological replicates). i, The distribution of AB352916 between the nucleus and cytoplasm analyzed using FISH. Scale bar: 5 μm. This experiment was repeated three times independently. Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with (a, c) or without Welch’s correction (a, f, g), one-way ANOVA followed by Tukey’s Multiple Comparison tests (b). ago-NC, negative control for agomir; anta-NC, negative control for antagomir.
Extended Data Fig. 3 The effects of CRAPIR knockout on the proliferation of cardiomyocytes.
a, The schematic for the construction of CRAPIR KO mice. b and c, Representative P1 and P7 heart tissue sections stained with AuroraB, and the quantification of the percentage of AuroraB+ cardiomyocytes (n=3 mice). White arrows point the positive cardiomyocytes. Scale bar: 20 μm. Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with Welch’s correction (c) or without Welch’s correction (b, c).
Extended Data Fig. 4 CRAPIR promotes cardiomyocyte proliferation and improves cardiac functions in P7 and adult mice with MI.
a, Masson staining of CKI mice subjected to MI. Scale bar: 500 μm. b, Cardiac sections from P7 mouse hearts stained with AuroraB at 3 days after MI (n=3 mice). Scale bar: 20 μm. c, The fibrosis of adult mice after MI for one month was studied using Masson staining (n=7 mice). Scale bar: 500 μm. d and e, Cardiac sections from adult mouse hearts stained with pH3 and Ki67 at 3 days after MI (n=3 mice). Scale bar: 50 μm. f, Cardiac sections from adult mouse hearts stained with AuroraB at 3 days after MI (n=3 mice). Scale bar: 20 μm. g, Echocardiography was used to observe cardiac function of adult mice at 28 days after MI (n=6 mice). Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with (e, f) or without Welch’s correction (b-g). MI, myocardial infarction; ago, CRAPIR agomir; ago-NC, negative control for agomir.
Extended Data Fig. 5 The expression of PA2G4 and the proteins binding to PA2G4.
a and b, The mRNA and protein expression of PA2G4 in cardiomyocytes transfected with PA2G4 siRNA and plasmid (n=5, 6 biological replicates). c and d, The protein expression level of PA2G4 in heart tissues (n=4 mice for c, 5 mice for d). e, Venn diagram showing overlapping proteins bind to PA2G4. Proteins with PSM values exceeding 5, unique peptide values greater than 10, and decreased abundance in PA2G4-bound proteins post-CRAPIR overexpression, which falls below 0.2 times that of the control group, were identified and selected. f, The structure of ILF2 and ILF3 proteins. g, Secondary mass spectrum for ILF2 and ILF3. Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with (a) or without Welch’s correction (a-d). AR, apex resection; NC siRNA, negative control for siRNA; OE, overexpression.
Extended Data Fig. 6 The effects of NF110 overexpression on cardiomyocyte proliferation and the influence among NF45, NF110, and PA2G4.
a, NF110 overexpression promotes cardiomyocyte proliferation (n=3 independent experiment). b, The effects of PA2G4 siRNA on the expression of NF90 and TCP80 (n=8 independent experiment). c, The effects of PA2G4 overexpression on the expression of NF110 in cardiomyocytes. This experiment was repeated three times independently. d, The quantification of Western blot results indicating the effects of PA2G4 on the expression of NF110 and NF45 in the nucleus and cytoplasm in cardiomyocytes (n≥4 independent experiment). e and f, The effects of NF110 and NF45 siRNAs on the protein expression level of PA2G4, NF110, and NF45 in cardiomyocytes (n≥5 independent experiment). g, The inhibitory effect of PA2G4 overexpression on the expression of NF110 was blocked by MG132 in cardiomyocytes. This experiment was repeated three times independently. h, Secondary mass spectrum for RNF13. Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with (b, d, e) or without Welch’s correction (a, b, d-f). NC siRNA, negative control for siRNA; OE, overexpression.
Extended Data Fig. 7 The effects of CRAPIR on the expression of NF110 and NF45 in cardiomyocytes and knockdown of NF45 attenuates the effects of CRAPIR on cardiomyocyte proliferation.
a, The protein expression level of NF110 and NF45 in cardiomyocytes transfected with CRAPIR (n≥6 independent experiment). b, The effects of CRAPIR on the expression of NF110 and NF45 in the nucleus and cytoplasm analyzed by Western blot. This experiment was repeated three times independently. c, The proliferative ability of cardiomyocytes was analyzed by the detection of EdU, pH3 and Ki67 (n=3 independent experiment). d, Co-IP analysis revealing the ubiquitination of NF110 was induced by CRAPIR antagomir in cardiomyocytes. This experiment was repeated three times independently. Data are presented as mean ± s.e.m. Statistical analyses were performed by Two-tailed Student’s t test with or without Welch’s correction (a), one-way ANOVA followed by Tukey’s Multiple Comparison tests (c). NC siRNA, negative control for siRNA; ago, CRAPIR agomir; ago-NC, negative control for agomir; anta, CRAPIR antagomir; anta-NC, negative control for antagomir.
Extended Data Fig. 8 Silencing of NF110 inhibits the improvement of CRAPIR on heart function after MI.
a, The heart contractile function was analyzed using Echocardiographic analysis (n=6 mice). b, The quantification of the percentage of pH3+ and Ki67+ cardiomyocytes (n=3 mice). c, The expression of cell cycle genes (n=4 biological replicates). Data are presented as mean ± s.e.m. Statistical analyses were performed by one-way ANOVA followed by Tukey’s Multiple Comparison tests (a-c). NC siRNA, negative control for siRNA; ago-CRAPIR, CRAPIR agomir; ago-NC, negative control for agomir; MI, myocardial infarction.
Extended Data Fig. 9 The effects of PA2G4 mutants on cardiomyocyte proliferation.
The proliferative ability of cardiomyocytes was analyzed by the detection of pH3 and Ki67 (n=3 independent experiment). Data are presented as mean ± s.e.m. Statistical analyses were performed by one-way ANOVA followed by Tukey’s Multiple Comparison tests.
Extended Data Fig. 10 The schematic of CRAPIR function in cardiomyocyte proliferation.
CRAPIR directly binds to PA2G4, affecting the interaction of PA2G4 with NF110-NF45 heterodimer and recruiting the ubiquitin ligase RNF13, reducing the degradation of NF110, thus promoting cardiomyocyte proliferation.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables 1–4
Supplementary Table 1 The proteomic data of CRAPIR pulldown. Supplementary Table 2 The quantification of western blot results indicating the binding of PA2G4 to NF110 and NF45. Supplementary Table 3 The quantification of western blot results indicating the binding of ILF3 to PA2G4 and NF45. Supplementary Table 4 The quantification of western blot results indicating the binding of NF45 to NF110 and PA2G4.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig./Table 1
Statistical source data.
Source Data Extended Data Fig./Table 2
Statistical source data.
Source Data Extended Data Fig./Table 3
Statistical source data.
Source Data Extended Data Fig./Table 4
Statistical source data.
Source Data Extended Data Fig./Table 5
Statistical source data.
Source Data Extended Data Fig./Table 6
Statistical source data.
Source Data Extended Data Fig./Table 7
Statistical source data.
Source Data Extended Data Fig./Table 8
Statistical source data.
Source Data Extended Data Fig./Table 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, W., Chen, H., Tian, Y. et al. The highly conserved PIWI-interacting RNA CRAPIR antagonizes PA2G4-mediated NF110–NF45 disassembly to promote heart regeneration in mice. Nat Cardiovasc Res 4, 102–118 (2025). https://doi.org/10.1038/s44161-024-00592-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-024-00592-z
This article is cited by
-
The PIWI-interacting RNA CRAPIR alleviates myocardial ischemia‒reperfusion injury by reducing p53-mediated apoptosis via binding to SRSF1
Acta Pharmacologica Sinica (2025)
-
Powering up piRNAs for heart regeneration
Nature Cardiovascular Research (2025)
-
Molecular gatekeepers of endogenous adult mammalian cardiomyocyte proliferation
Nature Reviews Cardiology (2025)
-
Stem-Cell Derived Exosomal microRNAs as Biomarkers and Therapeutics for Pediatric Cardiovascular Disease
Current Treatment Options in Cardiovascular Medicine (2025)