Abstract
The interaction between RAD51 and BRCA2 plays a key role in homologous recombination (HR), a critical DNA repair mechanism essential for the survival of cancer cells. Disrupting this interaction increases the sensitivity of cancer cells to chemotherapeutic agents. Here, we employed in silico methods to design aptamers—customized single-stranded oligonucleotides—specifically engineered to bind RAD51. These aptamers were developed with the aim of selectively modulating RAD51’s nuclear recruitment and its role in DNA repair processes. The leading candidate displays high affinity for RAD51, competing with BRCA2 for the same interaction site in vitro, as confirmed through biolayer interferometry (BLI) and fluorescence lifetime imaging microscopy (FLIM). In pancreatic cancer cells, we show that the aptamer impairs HR by altering the stress-induced nuclear localization of RAD51 and BRCA2, thereby reducing DNA repair efficiency and promoting the accumulation of DNA damage. Notably, when combined with the PARP inhibitor olaparib, the aptamer triggers synthetic lethality (SL) in a dose-dependent manner, an effect that is also preserved in 3D spheroid models. Our study showcases an aptamer-based approach for selectively targeting protein interactions within DNA repair pathways, introducing a promising avenue for SL-based treatments applicable to a wide range of cancers.
Similar content being viewed by others
Introduction
Cell growth and division inevitably cause DNA damage, which is normally resolved by dedicated repair pathways1. In cancer cells, rapid proliferation increases the likelihood of damage accumulation, leading to frequent activation of repair mechanisms2,3,4. Over time, sustained growth drives cancer cells to become highly proficient at engaging these pathways2,3. The specific pathway recruited depends on the nature of the lesion, such as strand breaks, base loss, or deamination5.
Double-strand DNA breaks (DSBs) are among the most deleterious forms of damage. Their repair primarily relies on homologous recombination (HR) or non-homologous end joining (NHEJ)6. The choice between these pathways, dictated by the cell cycle stage and the nature of the break7, is critical for preserving genomic stability and ensuring cell viability.
HR is a high-fidelity repair pathway that uses a homologous DNA sequence—most often the sister chromatid—to restore genome integrity. At the core of this process is RAD51, a key protein that mediates strand invasion and homology search8,9. RAD51 assembles on single-stranded DNA into nucleoprotein filaments that are critical for homologous recombination and DNA repair10,11. Its conformational flexibility underlies RAD51’s pivotal role in the accurate resolution of DSBs10.
Upon DNA damage, RAD51 is recruited to the nucleus by BRCA2, primarily through its interaction with eight highly homologous BRC repeats within BRCA2. Beyond recruitment, BRCA2 ensures the proper loading of RAD51 onto single-stranded DNA at the break site, enabling nucleoprotein filament formation12,13,14. BRCA2 also facilitates filament disassembly once repair is complete. The RAD51–BRCA2 interaction is therefore essential for the correct execution of homologous recombination. Targeting this complex has attracted significant attention in drug discovery, as its disruption could impair DNA repair—a vulnerability particularly critical in cancer cells that depend heavily on HR for survival. Although a high-resolution structure of the full RAD51–BRCA2 complex is not yet available, the high-affinity interaction between RAD51 and BRC4, the fourth BRC repeat of BRCA2, has been extensively characterized. BRC4 binds two specific pockets on RAD5115,16: the first pocket, which accommodates the FXXA motif, corresponds to residues Ala157–Met210, while the second pocket, which interacts with the LFDE motif, involves residues Asn196–Ser214 and Ser239–Gly26017,18,19. Molecules that selectively target this interface could sensitize cancer cells to DNA-damaging agents, thereby enhancing the efficacy of combined treatments20,21.
The combination of anticancer drugs is a widely adopted strategy to improve therapeutic outcomes, often exploiting the principle of synthetic lethality (SL)22. SL occurs when the simultaneous disruption of two genes or pathways results in cell death, whereas perturbation of either component alone is insufficient to induce lethality23. In BRCA-mutated cancers, where homologous recombination (HR) repair is compromised, SL can be achieved with poly(ADP-ribose) polymerase (PARP) inhibitors such as olaparib24, which block the completion of DNA repair through the base excision repair (BER) pathway25. Extending this rationale, the combination of PARP inhibitors with HR inhibitors in BRCA-proficient tumors sensitizes cancer cells to treatment regardless of their genetic background26,27. This dual-targeting strategy—conceptually aligned with what we have defined as “fully chemically-induced SL”28 —leverages common DNA repair dependencies and broadens the applicability of synthetic lethality, offering a promising avenue to improve therapeutic efficacy across cancers with high unmet medical needs. As such, it represents a paradigm shift in oncology, with the potential to transform treatment strategies and significantly improve patient outcomes29,30.
In the literature, small organic molecules, peptides, and peptidomimetics have been designed to target the RAD51–BRCA2 interaction21,31,32,33. However, none of these candidates have progressed to clinical development, and, to the best of our knowledge, no active clinical programs currently focus on compounds disrupting this interface. This underscores the complexity of disrupting BRCA2/RAD51 interaction, pinpointing the urgent need to develop new, potent molecules endowed with this mechanism of action and able to trigger synthetic lethality. This work investigates the use of aptamers as a strategy to disrupt the RAD51–BRCA2 interaction, thereby impairing HR within the framework of synthetic lethality. Aptamers are short, single-stranded oligonucleotides that bind specific targets with high affinity and selectivity through their unique three-dimensional structures and tailor-made sequences34,35,36. Compared with conventional small molecules or antibodies, aptamers offer several advantages: they can be synthesized at high yield and low cost, are readily amenable to chemical modification to enhance stability and specificity37, and exhibit low immunogenicity, making them generally well tolerated38. Their compact size and defined structure also facilitate penetration into tissues that are often less accessible to larger biomolecules39. Aptamers have already found applications in therapeutic, diagnostic, and research settings, and their versatility positions them as promising candidates for next-generation biomedical strategies, particularly in targeted drug delivery and personalized medicine36,40.
To date, the predominant method for aptamer generation has been the Systematic Evolution of Ligands by Exponential Enrichment (SELEX), a laboratory-based procedure that relies on iterative cycles of selection and amplification of candidate sequences34,41. While SELEX can yield aptamers with high affinity for target proteins, it remains time-consuming, labor-intensive, and often limited in producing highly specific oligonucleotides. Recent progress in computational approaches has begun to overcome these limitations. In particular, we and others have demonstrated the power of in silico aptamer design, including the generation of de novo RNA aptamers against TDP-43, a protein implicated in neurodegeneration, which highlights the precision and efficacy of algorithm-driven strategies40,42. In this work, we applied our in-house algorithm catRAPID43,44, a computational tool that predicts protein–nucleic acid interaction propensities by integrating secondary structure, hydrogen bonding, and van der Waals contributions45,46, to design aptamers specifically targeting the RAD51–BRCA2 interaction. Previous efforts had already explored aptamers as inhibitors of RAD51. For example, Martínez et al. identified three SELEX-derived aptamers able to interfere with RAD51–ssDNA complex formation, but their dissociation efficiency was limited, restricting their potential for therapeutic application47. By contrast, the sequences described here were rationally designed to selectively disrupt RAD51–BRCA2 binding, a distinct and clinically relevant interaction at the core of homologous recombination.
Our results show that these in silico–generated aptamers not only bind RAD51 with high specificity but also impair its functional interaction with BRCA2, thereby reducing HR activity and synergizing with PARP inhibition within a synthetic lethality framework. Both biophysical and cellular data strongly support their activity, positioning these aptamers as a promising next-generation strategy for the development of nucleic acid–based therapeutics in cancer treatment.
Results
In silico-designed aptamers bind a specific region of RAD51
We developed an in silico pipeline to design aptamers that selectively bind to specific regions of RAD51. To interfere with RAD51 interactions with BRCA2, we focused on two key areas around the RAD51-BRC4 interaction pockets, FXXA and LFDE, which we named Region 1 and Region 2 (Supplementary Fig. 1A, B). Additionally, we included a third area, known as the helix-hairpin-helix (HhH) motif, involved in non-sequence-specific DNA binding. This part, designated as Region 3, serves as an internal control to evaluate whether the binding in Region 1 or Region 2 is stronger (Supplementary Fig. 1C). The catRAPID algorithm was used to design aptamers of 9-16 nucleotides in length43,48. This falls within the range of 10–20 nucleotides that we previously exploited to successfully design aptamers (Table 1)40. Indeed, by limiting aptamers’ length, we aim to ensure direct interaction with the binding interface. Longer structures can indeed expose additional sites for unintended protein interactions, increasing the risk of off-target effects49,50.
For all resulting sequences, an interaction score with each of the defined RAD51 regions was predicted, and the aptamers were ranked based on these scores, with the condition that the values were higher than those of a negative protein control, actin B. The results consist of a series of aptamers characterized by repetitive short motifs rich in G (Table 1 and Supplementary Table 1). When comparing the catRAPID scores relative to the predicted interaction propensities, all of the generated sequences display a higher value for Region 2 compared to Region 1 or Region 3 (Supplementary Table 1). According to the predictions, the aptamers generated with catRAPID would preferentially bind Region 2 of RAD51, thus specifically disturbing the interaction with BRC4 on the pocket LFDE. As a negative control, the interaction scores for rcApt—the reverse complementary sequences of the top-scoring aptamer (Apt1)— was also included and the scores resulted to be close to 0.
Interaction studies validate the predicted binding of selected aptamers towards RAD51
The predicted binding propensities between the aptamers with an interaction score for Region 2 > 8 and purified RAD51 were validated (Supplementary Fig. 3). Apt1 reverse complementary sequence—rcApt—was used as a negative control. The binding affinities of the aptamers were determined by identifying their dissociation constants (Kds) using biolayer interferometry (BLI): the stronger the interaction, the lower the Kd (Fig. 1). To ensure a tight binding towards the target protein, the aim was to identify those aptamer sequences with a Kd for RAD51 < 1 µM.
A BLI analysis illustrating the binding kinetics of the first 6 aptamers (Apt1 to Apt6) and a negative control (rcApt) with RAD51 protein at 25 °C. Each curve represents the interaction profile of an individual aptamer, from which their respective binding affinities towards RAD51 can be extrapolated. Spheres: experimental points; line: curve fitting. The curve relative to rcApt is reported as raw data, since no binding response was registered. Three replicates were performed, and the mean and standard deviation were calculated for each point within the binding curves. Below each curve, the dissociation constant (Kd) is indicated. (ND = not detectable). B A scatter plot showing the correlation between experimentally determined Kds of the aptamers with RAD51 and their affinities predicted by the catRAPID algorithm for Region 2. Source data are provided as a Source Data file.
The highest catRAPID interaction scores, 12.47 and 12.44, respectively, were found for Apt1 and Apt2, which exhibited the lowest determined Kds of 116 ± 14 nM and 138 ± 4 nM (Fig. 1A). Kds of 188 ± 13 nM and 413 ± 11 nM were displayed by Apt3 and Apt4, both associated with a catRAPID score of around 10. Kds of 936 ± 39 nM and exceeding 2 µM were presented by Apt 5 and 6, both assigned catRAPID interaction scores below 9 (Fig. 1A). As predicted, the negative control rcApt exhibited no binding (Fig. 1A).
The results of these analyses indicate that the sequences of Apt1-Apt6, predicted by catRAPID to interact strongly with RAD51, displayed a trend where a higher catRAPID score correlated with lower Kd values (Fig. 1A). A notably robust negative correlation of −0.85 was uncovered, thereby confirming that the affinity trend is in accordance with the catRAPID score and that the algorithm was able to correctly predict the aptamers with the highest binding probability (Fig. 1B).
An in silico-designed aptamer can disrupt RAD51 filaments in vitro
Following the prediction of binding propensities between the top 6 aptamer candidates and RAD51, their effect on the protein oligomerization state was investigated. Apt1 was selected as the aptamer with the highest affinity based on computational predictions and in vitro evaluations.
The impact of Apt1 on RAD51 oligomerization was assessed using negative staining transmission electron microscopy (TEM), incubating RAD51 with increasing concentrations of Apt1 (Fig. 2 and Supplementary Fig. 4A, B). In agreement with the literature, also in this study RAD51 in isolation forms mainly worm-like helical fibrils (Fig. 2A)31. Upon incubation of RAD51 with equimolar concentrations of Apt1, no visible effect was observed (Fig. 2B). However, increasing the concentration of Apt1 to six times that of RAD51 resulted in the almost total disruption of the long worm-like fibrils that were replaced by smaller oligomers (Fig. 2C). The resulting ring-like arrangements of the oligomers displayed an average diameter of 13.3 ± 1.4 nm (Supplementary Table 2). A further increase in the concentration of Apt1 to twelve times that of RAD51 led to the complete disruption of the oligomers (Fig. 2D), indicating that the aptamer significantly affected RAD51 quaternary structure, particularly in terms of filament stability. Incubating RAD51 with the highest concentration of the negative control aptamer, rcApt, did not perturb the filamentous structure of the protein (Supplementary Fig. 4C).
A RAD51 by itself, displaying the characteristic fibril structure. B RAD51 incubated with Apt1 at a molar ratio of 1:1, exhibiting similar supramolecular structures. C RAD51 incubated with Apt1 at a molar ratio of 1:6, revealing a significant reduction in the presence of long fibrils, accompanied by the formation of smaller oligomeric structures. D RAD51 incubated with Apt1 at a molar ratio of 1:12, resulting in the complete abrogation of fibrils. Experiments were performed in biological triplicate, and data were collected from eight images per experiment. Source data are provided as a Source Data file.
These results indicate an interference exerted selectively by Apt1—and not by rcApt—on the oligomerization state of RAD51. By interfering with RAD51 quaternary structure, the aptamer might also affect the protein’s ability to repair damaged DNA via HR.
Computational predictions confirm the structural alterations in RAD51 filaments induced by APT1
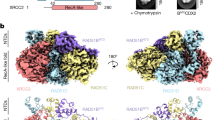
To gain atomistic details on the mechanisms by which Apt1 disassembles RAD51 filaments, structural predictions of RAD51 fibrils in the presence of increasing copy number of Apt1 (1, 6 or 12) were performed using AlphaFold 351 (Fig. 3). As a control, the same predictions were repeated with rcApt (Supplementary Fig. 5).
A RAD51 by itself (6 protomers), displaying the characteristic fibril structure. B One filament of RAD51 in complex with one Apt1 molecule, exhibiting a similar supramolecular structure with A. C One filament of RAD51 in complex with six Apt1 molecules, revealing a reorganization of the protofilament into a hexameric annular structure. D One filament of RAD51 in complex with 12 Apt1 molecules, resulting in the complete abrogation of the fibrils. In this latest case, two trimers are interacting with several copies of the aptamer. Each protein chain is shown in a different color using the cartoon-based representation (chain A in orange, B in magenta, C in cyan, D in pink, E in silver and F in green). Each aptamer copy is reported as a blue ribbon, with the sugar rings in yellow and the nucleobases in red.
Following up on one of the resolved structures of a RAD51 filament in which one helical turn corresponds to 6.6–7.3 RAD51 protomers per turn52, the prediction of the RAD51 filament was attempted with both six and seven monomers. It was found that the former was better suited to obtain a model (Fig. 3A) that closely resembled the X-ray helical structure of RAD51 in the absence of the ssDNA filament to be repaired, defined as apo protein (PDB ID: 5NWL)52. A relevant difference between the deposited structure and the model here proposed is the reconstruction of the unresolved loop from Ala271 to Gly288. This portion (part of L2 in Pellegrini et al.19 connecting β-strand B5 to β-strand B6) was crucial to properly model the interaction with the aptamers53. The prediction of the structure including 7 RAD51 copies was perfectly overlapping with the one containing 6 copies, with the seventh protomer beginning a new helical turn, and it is not reported to avoid redundancy.
The model here proposed is characterized by a pitch of 125 Å, fitting the previously reported range of 112–134 Å52 for the apo protein. This reconstruction of RAD51 fibril structure was used as a reference to evaluate the predicted complexes formed by the protein filament with increasing copies of Apt1 molecules.
In the model with one copy of Apt1, the aptamer interacts with five of the six RAD51 molecules forming the filament, without impacting its structure (Fig. 3B). In accordance with the design, the aptamer is located in a different groove (Supplementary Fig. 2B) compared to the protein’s ssDNA binding site identified by cryoEM (PDB ID: 8PBC)54. In particular, the aptamer lies juxtaposed to the DNA binding site used for repair, sharing with it the interaction with the backbone atoms of a few amino acids only (Supplementary Table 3). In this configuration, the pitch length shrunk to 115 Å, still in accordance with the deposited apo structure (second assembly of the 5NWL PDB)52. In this framework, 17 out of 25 predicted models were found binding Apt1 at a different spot than the RAD51 ssDNA binding site (Fig. 3B); in other 5 configurations, Apt1 was bound to the ssDNA binding site; in 3 out of 25 models, Apt1 was binding the filament at different spots with weak and non-specific interaction, or not binding at all.
To interpret the TEM results, the effect of 6 copies of Apt1 on the disassembly of one RAD51 filament was predicted (Fig. 3C). The resulting model revealed the loss of the helical turn of RAD51 filament and the rearrangement of the six RAD51 protomers into a circular complex, with each of the aptamer molecules intercalating between adjacent chains. This peculiar structure for a RAD51 oligomer displayed an approximate diameter of 13 nm, qualitatively resembling the smaller non-filamentary oligomeric structures identified by TEM imaging at the 1:6 molar ratio (Fig. 3C). According to the structural predictions, the amino acids of RAD51 filament involved in the interaction with the aptamer only partially overlapped with the ones found in the model comprising one RAD51 filament and one Apt1 molecule (Supplementary Table 3): among them, Arg229, Arg235, Leu238, Ser239, Ala271 and Asn290. Conversely, the unique intermolecular interactions borne by the annular-shaped complex involved some of the residues of the reconstructed loop: Phe279, Ala280, Ala281, Asp282 and Pro283. Statistically, the proposed circular oligomeric structure was observed in 15 out of 25 predictions. Conversely, in only 5 out of 25 predictions, the RAD51 filament was able to maintain its structure in the presence of six copies of Apt1. Among these, five showed non-specific interactions with the filament, and only one bound to the ssDNA binding site. In the remaining 5 predictions, the oligomeric structure was disrupted, yielding a mix of dimers and trimers connected by aptamers.
Recapitulating the TEM condition with the highest amount of aptamer, RAD51 filament was then modeled with 12 Apt1 molecules. None of the AlphaFold 3 generated structures resulted in a helical filament, nor in a circular structure. Instead, the filament is broken in two parts, resulting in two trimers interconnected with the aptamers, mostly in a non-specific arrangement (Fig. 3D). Indeed, nine out of 12 aptamer strands were only bound to the proteins by means of non-specific and non-conserved contacts, lying between the two trimers and establishing occasional interchain base pairs with other aptamer copies. There are two exceptions: i) two Apt1 molecules binding the RAD51 protomers (Fig. 3D) at a similar spot as in the one filament-one aptamer model (Fig. 3B), mostly via the same amino acids (Supplementary Table 3), albeit with a different binding mode; ii) another aptamer contacting the protein between chain E and F (in silver and green, respectively, in Fig. 3D), close to Asp274-Pro283.
Both the target patches involved in the interaction with the nucleotides of Apt1 are part of the Region 2 of RAD51, the target area for which the aptamers were specifically designed (Supplementary Figs. 1B and 2 and Supplementary Table 3). Instead, in contrast with Apt1, the structural predictions in the presence of an increasing copy number of rcApt show that also in silico the control aptamer is not able to interfere with the RAD51 filament formation (Supplementary Fig. 5).
The aptamer affects BRC4–RAD51 interaction
To rigorously assess the binding specificity of the designed aptamers to the targeted pockets on RAD51, Fluorescence Lifetime Imaging Microscopy (FLIM) was employed. This technique enables us to probe molecular interactions with high precision by measuring changes in fluorescence lifetimes, which can indicate binding events at a molecular level55,56. Here, FLIM was used to investigate whether the aptamers could effectively influence the interaction between RAD51 and the BRC4 peptide.
The best performing aptamer, Apt1, was selected to be studied with this technique and labeled with Texas Red (Fig. 4). As a control, rcApt, which does not bind RAD51 (Fig. 4), was included.
Phasor plot from FLIM analysis showing sine component (S) and cosine component (G) on the axes, for the fluorophore Texas Red conjugated to the aptamers Apt1 and rcApt. Points A–F correspond to different experimental conditions. A Apt1 alone. B Apt1 after incubation with RAD51. C Apt1 with RAD51 followed by the addition of BRC4. D rcApt alone. E rcApt after incubation with RAD51. F rcApt with RAD51 followed by the addition of BRC4. D–F Results are perfectly overlapping. Source data are provided as a Source Data file.
The FLIM experiments were performed in various conditions: the lifetime fluorescence of Texas Red directly conjugated to Apt1 and to rcApt was measured alone, in the presence of RAD51, and in the presence of RAD51 together with BRC4. The fluorescence lifetime decay of Apt1 was analyzed before (Fig. 4, point A) and after (Fig. 4, point B) the addition of RAD51, revealing an increase in the slow component (τslow) from 5.01 ± 0.1 ns to 5.32 ± 0.2 ns (Table 2). This increase of τslow indicates an interaction between RAD51 and Apt1. Introducing BRC4 into this system (Fig. 4, point C) led to a reduction in τslow down to 5.09 ns, suggesting a partial displacement of Apt1 from its binding site by BRC4, thus reintroducing the aptamer into solution. The observed intermediate lifetime of 5.09 ± 0.1 ns supports the theory that Apt1 was both in solution and partially bound to RAD51, indicating that its interaction with RAD51 affects the protein’s capacity to bind BRCA2.
Furthermore, the decomposition of the fluorescence signal into ratios of “slow” and “fast” components revealed the presence of multiple fluorescent states, each of which differentially influences the fluorescence lifetime. The fast component, indicating a short lifetime, suggests rapid return to the ground state, whereas the slow component, indicating a longer lifetime, suggests a slower return. The increase in the Apt1’s slow component from 0.71 to 0.82 upon adding RAD51, and its subsequent return to 0.71 with the addition of BRC4, supports that BRC4 and Apt1 both target a common region of RAD51 (Table 2). Predicted complexes from AlphaFold 3 showing the positioning of BRC4 and Apt1 at their respective binding sites on the RAD51 filament are presented in Supplementary Fig. 6, along with the model of the RAD51 oligomer in the presence of excess BRC4 peptide (Supplementary Fig. 7). A parallel experiment with rcApt consistently showed a constant τslow of 5.01 ± 0.1 ns under all tested conditions: alone, in the presence of RAD51, and in the presence of RAD51 and BRC4 (Fig. 4, points D, E, F). This uniformity in decay time irrespective of the presence of RAD51 or BRC4 across these conditions confirms that the negative control rcApt does not interact with either molecules.
To further validate the results obtained by FLIM, BLI was also employed (Supplementary Fig. 8). BLI experiments were conducted by immobilizing biotinylated Apt1 onto streptavidin-coated biosensors and measuring RAD51 binding in the absence or presence of increasing concentrations of BRC4. In the absence of BRC4, the RAD51–Apt1 interaction produced a strong binding response. The addition of increasing concentrations of BRC4 progressively reduced the RAD51–Apt1 signal, indicating clear competition between the aptamer and the peptide.
These findings provide strong evidence that the designed aptamers are interacting with the intended regions on RAD51, validating the in silico predictions and confirming that the aptamers have the potential to disrupt the RAD51-BRCA2 interaction, offering a promising strategy for impairing the homologous recombination pathway in cancer cells.
The aptamer alters the cellular localization of RAD51 and BRCA2
The aptamers’ effects against RAD51 activity in a cellular environment were assessed in pancreatic ductal adenocarcinoma (PDAC) BxPC-3 cells. This cell line was selected as a BRCA2-proficient PDAC model in our experiments for its high RAD51 expression, absence of BRCA2 mutations, and presence of mutations in the classic PDAC driver genes TP53 (Y220C, missense variant), CDKN2A (gene deletion) and SMAD4 (gene deletion)57. The aptamer sequences were stabilized against nuclease activity with the introduction of locked bases. Apt1, the best-performing aptamer, and the negative control rcApt were selected for further investigations.
Under physiological conditions, RAD51 is distributed mostly in the cytoplasm. To participate in DNA repair via HR, RAD51 is recruited from the cytosol into the nucleus and brought to the damage site by BRCA258,59. Upon recruitment to the nucleus in response to DSBs, both RAD51 and BRCA2 accumulate at the damage site, forming distinctive nuclear foci, regions of DNA where the damage occurs60. One way to investigate the effect of the aptamers on the BRCA2-driven RAD51 nuclear recruitment is to evaluate the subcellular localization of the proteins under stress. To recreate stressed conditions, cisplatin was employed. Cisplatin is a double DNA breakage compound that forms covalent bonds with DNA bases, leading to the creation of adducts, which can distort DNA’s structure61,62.
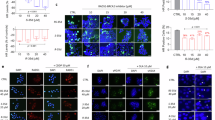
Laser scanning microscopy with Single-Photon Avalanche Diode (SPAD) was employed to assess the subcellular localization of RAD51 and BRCA2 under DNA damage conditions63, in the presence or absence of increasing concentrations of Apt1 (Fig. 5 and Supplementary Fig. 9). Under unstressed (physiological) conditions, RAD51 was primarily cytoplasmatic, while BRCA2 showed a diffuse distribution across the cytoplasm and nucleus (Fig. 5A). Following cisplatin treatment, both proteins accumulated in the nucleus (Fig. 5A), consistent with their roles in homologous recombination repair64. However, co-treatment with Apt1 impaired this nuclear relocalization in a dose-dependent manner (Fig. 5A). At 25 μM Apt1, protein distribution was unaffected; 50 μM induced minor, non-significant changes (Fig. 5A). 100 μM and 200 μM Apt1 markedly reduced RAD51 nuclear accumulation and prevented BRCA2 nuclear enrichment, restoring a distribution resembling unstressed conditions (Fig. 5A). Pearson’s correlation analysis confirmed the statistical significance of this effect at 100 μM and 200 μM Apt1 (Supplementary Table 4). No impact was observed with the reverse complementary control rcApt (Supplementary Fig. 10, Supplementary Table 4). To further quantify these effects, image-based analysis classified nuclear fluorescence intensity (NFI) for RAD51 and BRCA2 (Fig. 5B). In untreated cells, RAD51 and BRCA2 NFIs averaged ~10% and ~5%, respectively (Fig. 5B). Cisplatin increased RAD51 NFI to ~50%, while BRCA2 showed two distinct populations centered at ~15% and ~35% (mean ~20%) (Fig. 5B). Apt1 at 25 μM or 50 μM did not significantly alter these distributions, though BRCA2’s bimodal pattern was lost at 50 μM (Fig. 5B). At 100 μM Apt1, RAD51 NFI dropped to ~15%, and BRCA2 shifted to ~35% (Fig. 5B). At 200 μM, both proteins’ NFIs approximated untreated levels (RAD51 ~ 5%; BRCA2 ~ 43%, (Fig. 5B).
A High-resolution immunofluorescence images of a single BxPC-3 cell untreated or treated with increasing concentrations of Apt1 in combination with cisplatin. Nuclei are stained with DAPI (blue); RAD51 is shown in green; BRCA2 in red. Scale bar: 10 μm. B Bar graphs illustrating the image-based quantification of a cell population nuclear fluorescence, showing RAD51 (orange) and BRCA2 (blue) under DNA damage conditions and increasing concentrations of Apt1. Experiments were performed in biological triplicates, and data were collected from approximately 500 cells per condition. Source data are provided as a Source Data file.
Furthermore, to quantitatively assess the effect of Apt1 on RAD51 nuclear localization, we employed immunofluorescence microscopy. RAD51 nuclear fluorescence was measured under physiological conditions, under stress-induced conditions (cisplatin treatment), and following co-treatment with increasing concentrations of Apt1. Under physiological conditions, nuclear RAD51 fluorescence represented approximately 10%, increasing to 65% following cisplatin treatment (Supplementary Fig. 11). Co-treatment with 25 µM and 50 µM Apt1 resulted in moderate decreases to 56% and 42%, respectively. A more pronounced reduction was observed with 100 µM and 200 µM Apt1, lowering nuclear fluorescence to approximately 25% and 10%, respectively (Supplementary Fig. 11). In contrast, treatment with the negative control rcApt had no significant effect (Supplementary Fig. 10).
In addition, a pixel-based colocalization study of RAD51 and BRCA2 was conducted to gain further insights on Apt1’s mechanism of action at the molecular level (Supplementary Fig. 12 and Supplementary Table 5). For this purpose, 10 representative cells per condition were selected. The Manders’ coefficient (M) was employed to quantify the degree of colocalization between RAD51 and BRCA2 nuclear signals. By determining M, the fraction of RAD51 signal (green) overlapping with BRCA2 signal (red) at the pixel level was quantified. Values range from 0 (no colocalization) to 1 (complete colocalization). Under unstressed conditions, the NFI of both RAD51 and BRCA2 was minimal, resulting in an M value close to zero, consistent with the absence of both proteins in the nuclear compartment (Supplementary Table 5). Upon cisplatin treatment, RAD51 and BRCA2 accumulated in the nucleus to facilitate HR, reflected by an increased M value of 0.44, indicating that over 40% of nuclear RAD51 is found in close proximity to nuclear BRCA2 (Supplementary Table 5). Co-treatment with Apt1 resulted in a dose-dependent reduction of nuclear colocalization. Specifically, when combining cisplatin with 25 µM or 50 µM Apt1, M values were approximately 0.35 and 0.33, respectively, suggesting that this aptamer dose cannot affect RAD51 and BRCA2 nuclear colocalization (Supplementary Table 5). At higher concentrations (100 µM and 200 µM Apt1), M values were reduced to near zero, indicating a marked impairment of RAD51-BRCA2 colocalization (Supplementary Table 5). Treatment with 200 µM negative control rcApt had no observable effect, maintaining an M of 0.42, a result similar to that of cisplatin-only treated cells, thereby confirming the specificity of Apt1 action (Supplementary Table 5).
This analysis supports the hypothesis that Apt1 interferes with the nuclear colocalization of RAD51 and BRCA2 in a dose-dependent manner, potentially affecting HR-mediated DNA damage repair.
The aptamer impairs homologous recombination efficiency leading to DNA damage accumulation
To investigate the aptamers’ impact on RAD51-BRCA2-dependent HR, Apt1 and rcApt ability to affect HR efficiency was assessed at the single effective concentration of 100 nM in BxPC-3 cells, using the functional HR reporter assay “HR-Quick Assay” (HR-QA), also employed in our previous publications investigating RAD51-BRCA2 small molecule inhibitors21,31,57,65,66. HR-QA data show that, while rcApt did not affect HR efficiency compared to control cells (CTRL), Apt1 induced a significant and strong (50.5%) inhibition of HR activity (Fig. 6A), in line with the reduced Apt1-mediated RAD51 nuclear localization and further supporting Apt1 negative effect on HR activity. To corroborate HR-QA data, additional HR inhibition studies were performed with the mClover-Lamin A assay (mCL-HR). This assay exploits a HR-mediated insertion of an mClover-containing homologous sequence into a Cas-9-generated DSB in the Lamin A gene (LMNA). After the Cas-9-operated cleavage, the DNA DSB is repaired via HR with the mClover-containing homologous sequence, resulting in the reconstitution of a fluorescent mClover-Lamin A fusion protein. Since this assay requires highly transfection-permissive and proliferating cells, mCL-HR was performed on HEK293 (whose higher transfection efficiency was previously assessed66) instead of BxPC-3 cells, characterized by a lower transfection efficiency and a slower doubling time67,68. Consistently with HR-QA results, mCL-HR data show a significant dose-dependent (50-100-200 nM) effect of Apt1 on HR efficiency (50 nM: 80.9%; 100 nM: 51.2%; 200 nM: 38.9%) compared to control cells, while rcApt did not significantly impact HR activity in the same treatment conditions (Fig. 6B). These data emphasize the downstream effect of the interference that Apt1 exerts on RAD51-BRCA2 interaction, highlighting the potential of Apt1 as inhibitor of RAD51-BRCA2-dependent HR.
A Effect of Apt1 and rcApt (100 nM) on HR efficiency (measured via HR-QA) in BxPC-3 cells. Results are expressed as mean ± SD (n = 4). Statistical analysis was performed with one-way ANOVA followed by Dunnett’s multiple comparison test. B Analysis of HR-positive (mClover-Lamin A-positive) HEK293 cells after administration of increasing doses of Apt1 or rcApt (50, 100 and 200 nM. Results are expressed as mean ± SD (n = 4). Statistical analysis was performed with one-way ANOVA followed by Dunnett’s multiple comparison test, with **p < 0.01 and ****p < 0.0001. Experiments were performed in biological quadruplicates, and data were collected from ten images per condition. C Bar chart showing the percentage of BxPC-3 cells positive for γH2AX foci 72 h after transfection with Apt1 or rcApt, alone or in combination with 10 μM olaparib (OLAP). Results are expressed as mean ± SD (n = 3, 10 images per sample). Statistical analysis: two-way ANOVA followed by Tukey’s multiple comparison test; (§§§§p < 0.0001 vs CTRL; ####p < 0.0001 vs OLAP; ****p < 0.0001 vs aptamer alone). D, E Immunofluorescence images of BxPC-3 cells stained for γH2AX foci 72 h after transfection with aptamer alone (D) or co-treated with 10 μM olaparib 48 h (E). Insets highlight a cell positive for γH2AX foci. Scale bar: 50 µm. Experiments were performed in biological triplicates, and data were collected from ten images per condition. Source data are provided as a Source Data file.
If HR is impaired, cells cannot promptly repair DSBs. In particular, cancer cells, characterized by higher replication rates, heavily rely on HR-mediated DNA damage repair69. These pathophysiologic conditions, coupled to DNA quality control deficiencies (e.g., due to TP53 mutations) often observed in different tumor types, make cancer cells more susceptible to DNA damage accumulation70,71,72,73. In this regard, a prolonged HR inhibition results in increased DNA damage, further exacerbated upon the inhibition of PARP-mediated SSBs DNA repair mechanisms3. Therefore, to investigate the potential of the aptamers in halting DNA repair and amplifying drug-induced DNA damage accumulation, we analyzed DSB events by quantifying the number of foci formed by phosphorylated histone variant γH2AX74 using IF74 after exposure to the aptamers, alone or in combination with the drug olaparib. Olaparib, a PARP inhibitor (PARPi), is primarily used in cancer patients affected by mutations in the BRCA1 or BRCA2 genes24,25. These mutations impair the cells’ ability to repair DNA through the HR pathway, leading to increased reliance on alternative repair mechanisms, such as PARP-mediated base excision repair. By inhibiting PARP enzymes, olaparib exacerbates DNA damage accumulation in these already compromised cancer cells, ultimately leading to cell death. This single olaparib concentration of 10 μM was chosen to approximate clinically relevant olaparib plasma concentrations (Cmin = 1 μM, Cmax = 8.5 μM) reported in adult pancreatic cancer patients with BRCA2 germline mutations receiving olaparib 100mg BID in co-administration regimens with other drugs, such as gemcitabine24,75.
The baseline levels of DSBs were investigated in cells left to grow for three days (Fig. 6C–E). In the absence of aptamers, approximately 12% cells were positive for γH2AX foci (Fig. 6C–E). However, in the presence of Apt1 the cells positive to foci reached 21.1%, a significant increase compared to both cells untreated and treated with the negative control rcApt (10.9%, Fig. 6C–E). This result suggested that Apt1, but not rcApt, interfered with physiological repair processes.
IF microscopy was employed to assess the effect of Apt1 and of the negative control rcApt on γH2AX levels, in the presence of the PARP inhibitor olaparib. Only 14% of untreated cells were positive for γH2AX foci (Fig. 6C–E). Cells treated with olaparib and Apt1 showed a significant increase in γH2AX foci (54.2%) compared to cells treated solely with olaparib (30.9%) (Fig. 6E). In contrast, cells treated with olaparib and rcApt showed a negligible increase in γH2AX foci (35.3%) (Fig. 6C–E). The increment in the number of these species induced by Apt1 in combination with olaparib is indicative of the accumulation of DNA damage that escapes repair mechanisms. A concurrent and sustained inhibition of both DNA DSBs and SSBs repair ultimately leads to genomic instability, chromosome aberrations and the acquisition of mutations3. Therefore, the extent of genomic instability in BxPC-3 after the exposure for 24, 48 or 72 h to Apt1 or rcApt (100 nM), alone or in combination with 10 μM olaparib, was assessed by evaluating the presence of micronuclei, small DNA-staining bodies outside the main nucleus. In line with previous results on DNA damage, only Apt1 amplified olaparib-induced micronuclei formation at 48 h and further exacerbated after 72 h exposure, while rcApt was found ineffective (Supplementary Fig. 13). Altogether, these data are consistent with Apt1 ability—but not rcApt—to affect HR efficiency and to amplify olaparib-mediated DNA damage accumulation, further supporting its mechanism of action and suggesting a potential SL profile for Apt1.
The aptamer induces synthetic lethality in cancer cells in combination with OLAPARIB
Once established, the effect of the aptamers on physiological accumulation of γH2AX foci, their impact on cell viability was assessed to investigate aptamer/PARPi synergism and to confirm SL manifestation. To sensitize target cells to treatment by combining olaparib with an aptamer acting as HR inhibitor, BxPC-3 cells were exposed to increasing concentrations of Apt1 and rcApt (50, 100 and 200 nM), alone or in combination with 10 μM olaparib (0.1% DMSO as vehicle control, CTRL). Our results demonstrate that, while rcApt did not significantly impact cell viability at any tested concentration, Apt1 showed a clear dose-dependent reduction in viability (50 nM: 83.5%; 100 nM: 74.5%; 200 nM: 71.9%) and significantly amplified olaparib-mediated death (50 nM: 54.4%; 100 nM: 51.4%; 200 nM: 40.2%) in BxPC-3 cells (Fig. 7A). Drug interaction index (i.index) calculations were then performed to quantify the extent of potential synergism between aptamers and olaparib (as detailed in the Materials and Methods section). A synergistic effect was observed only for Apt1 at both 100 nM (i.index = 0.836 ± 0.050) and 200 nM (i.index = 0.719 ± 0.054), confirming our original hypothesis and the manifestation of a chemically induced SL. In parallel, to explore the selectivity of the aptamer/olaparib co-administration for cancer cells, the potential toxicity (cell viability <80% vs CTRL, as indicated by the red threshold) of the combination was assessed in the non-cancerous epithelial cell line MCF-10A, following the same treatment regimen employed for BxPC-3 cells. Our data show that both single agents (i.e. Apt1, rcApt and OLA) and the aptamer/olaparib combinations were unable to significantly affect cell viability at the same concentrations tested on BxPC-3 cells (Fig. 7B). These results are consistent with literature data indicating cancer cells’ overdependency on DNA damage response pathways and their consequent upregulation69. Highly proliferating cancer cells like BxPC-3 overexpress RAD51 and over-rely on RAD51-BRCA2-dependent HR69. This renders a RAD51-BRCA2/PARP inhibition-based SL strategy effective to selectively kill tumor cells. Conversely, non-cancerous cells like MCF-10A display a full complement of DNA repair pathways, mitigating the impact of RAD51-BRCA2/PARP inhibition on cell viability. This may be due to a higher compensation for the aptamer/PARPi-induced loss of individual DNA damage repair pathways76. These findings strengthen and extend our initial observations demonstrating that the combined Apt1/PARPi treatment significantly enhances SL in a dose-dependent manner only in cancer cells, while its cytotoxic effect on normal cells appears minimal.
A–C Cell viability after 72 h treatment with increasing Apt1 or rcApt doses (50, 100, 200 nM) alone or in combination with 10 μM olaparib (OLAP) in BxPC-3 cells (A), MCF-10A cells (B) or BxPC-3 3D spheroids (C). D–F Representative brightfield and live/dead staining images (D), time-course spheroid volume quantification (E) and cell death assessment (F) of BxPC-3 3D spheroids after 72 h treatment with increasing 200 nM Apt1 alone or in combination with OLAP. Results are expressed as mean ± SD. Experiments were performed in n = 5 biological replicates. Statistical analysis: two-way ANOVA followed by Tukey’s multiple comparison test, with §p < 0.05 or §§p < 0.01 vs CTRL; #p < 0.05, ##p < 0.01 or ###p < 0.001 vs OLAP; *p < 0.05 or **p < 0.01 vs aptamer alone. Source data are provided as a Source Data file.
This analysis provides critical insights into the optimal concentration ranges to maximize cancer cell death through dual inhibition of DNA repair pathways while concurrently limiting cytotoxicity in normal tissues. To gain further insight into the potential of our approach to effectively tackle PDAC and the possibility of Apt1 future development as a pharmacologic agent within the SL rationale, we also tested combinatorial effects in BxPC-3 3D spheroids, which better replicate in vivo tumor conditions of cell-cell and cell-matrix interactions57. Following our established protocol, we evaluated cell viability, spheroid volume, and cell death. Consistent with our 2D data, rcApt did not significantly impact cell viability in the 3D system, while Apt1 produced a dose-dependent reduction in viability when administered alone (50 nM: 85.2%; 100 nM: 83.1%; 200 nM: 79.5%) and further amplified olaparib-mediated cancer cell death (50 nM: 70.1%; 100 nM: 59.4%; 200 nM: 50.5%) (Fig. 7C). Notably, in the 3D setting, synergistic interaction was observed only at 200 nM Apt1 (i.index = 0,794 ± 0,039), likely reflecting differences in drug resistance and the more difficult accessibility of 3D spheroids compared to monolayer cultures77,78. To further evaluate Apt1 antineoplastic effects, we monitored changes in 3D spheroid volume during a time-course treatment with Apt1 at the dose effective in 3D cultures, i.e. 200 nM, alone or in combination with 10 μM olaparib (OLAP). In agreement with cell viability results, single agents slightly but significantly decreased spheroid volume (Apt1 200 nM: 81.8%; OLAP: 80.9%), which was further reduced by their association (Apt1 + OLAP: 69.9%) (Fig. 7D, E). Moreover, live/dead staining with vital dyes Calcein-AM and PI showed increased spheroid death upon treatment with either Apt1 (mean ratio = 0.70) or 10 μM olaparib (OLAP) (mean ratio = 0.74), which was amplified by their coadministration (mean ratio = 1.19) (Fig. 7F). These results reinforce our original rationale and the manifestation of a chemically induced SL occurring as a consequence of a dual inhibition of DNA repair pathways. Importantly, the consistent findings in both 2D and 3D models support the potential of Apt1 as a therapeutic agent, guiding future dose optimization strategies for preclinical development and offering a promising strategy to exploit vulnerabilities in DNA repair pathways for improved cancer treatment outcomes.
Discussion
The RAD51-BRCA2 interaction is central in HR, enabling an accurate DNA DSBs repair, and has recently emerged as an attractive target for cancer therapy. RAD51 requires BRCA2 for recruitment and filament assembly at sites of DNA damage, and disrupting this partnership compromises HR and sensitizes cells to DNA-damaging agents. Small molecules and peptides have validated this strategy, disrupting RAD51 foci formation and sensitizing cells to PARPi-mediated cell death18,31,33. Together, these studies underscored the therapeutic value of disrupting the RAD51-BRCA2 interaction as a compelling pharmacological approach in oncology.
Our computationally driven approach not only identified aptamers that specifically bind to the critical RAD51 interaction sites with BRCA2, but also provided a robust validation of their binding affinity and specificity. Structural and mechanistic studies revealed that Apt1 prevents RAD51 filament formation. TEM experiments and AlphaFold351 predictions show that Apt1 disrupts protomer–protomer interactions, shifting the equilibrium toward short oligomeric structures (e.g. trimers, hexamers) instead of extended stable filaments. FLIM results and BLI competition assay further indicate that Apt1 competes with BRC4 for the same binding site or that BRC4 induces conformational changes that prevent aptamer association. Thus, Apt1 functionally recreates a BRCA2-deficient state and destabilizes RAD51 quaternary structure, affecting its function to repair the damaged DNA via HR.
These mechanistic observations directly translate into marked effects in cellular contexts. High-resolution SPAD-based microscopy results indicate that Apt1, but not rcApt, interferes with the DNA damage-induced nuclear translocation of both RAD51 and BRCA2, likely preventing filament assembly63. This reduction of Apt1-induced RAD51 nuclear localization functionally disables BRCA2-dependent DNA repair and consistently impairs HR efficiency in a dose-dependent manner, leading to unrepaired DNA DSBs accumulation and micronuclei formation upon concurrent PARP inhibition. These findings support Apt1 specific pharmacological effect to reproduce a BRCA2-deficient state, highlighting the potential for fine-tuning aptamer dosing to achieve desired levels of DNA repair inhibition.
Importantly, by functionally mimicking BRCA2-mutated pathological phenotype and sensitizing the olaparib-mediated cell death, Apt1 enables a SL paradigm in BRCA-proficient PDAC cells. This expands the use of PARPi therapy beyond inherited BRCA mutations and directly translates the paradigm of “fully chemically-induced SL” from small organic molecules to aptamers, paving the way for the development of novel pharmacological approaches21,31,57,65,66. This is of particular interest given that only a small fraction of PDAC patients harbor BRCA1/2 mutations and thus benefit from PARPi therapy79. By inducing a BRCAness phenotype, Apt1 could broaden SL therapeutic reach, offering a strategy to sensitize a wider range of patients to PARPi and/or potentially overcome primary resistance mechanisms32. Notably, our aptamer/PARPi SL approach did not impair viability in non-cancerous epithelial cells. This differential activity underscores Apt1 tumor-selective nature of Apt1, consistent with its mechanism of exploiting elevated RAD51 expression and HR dependency in cancer cells80,81,82. Such selectivity is essential for minimizing toxicity in normal tissues and strengthens the translational potential of our approach. Moreover, by driving persistent DNA damage and micronuclei formation, the Apt1/olaparib combination may also enhance tumor immunogenicity by activating innate immune sensors such as cGAS–STING or RIG-I/MDA5, suggesting that our aptamer/PARPi SL approach could sensitize tumor cells to immunotherapies83. The possibility to expand SL beyond BRCAness and to potentiate immune responses further underscores the potential of Apt1 as a versatile anticancer tool.
Unlike small molecules, which often face limitations in targeting shallow protein–protein interaction surfaces, or peptides, which may suffer from stability and delivery issues, aptamers combine molecular precision with adaptable chemistry84. Their structural flexibility allows them to mimic key protein interaction motifs while offering the possibility of chemical modifications to improve half-life and delivery. This positions aptamers as a complementary modality that could bridge the gap between small-molecule druggability and the specificity of biologics. Nevertheless, aptamers’ high selectivity and reversibility make them powerful probes to study RAD51 filament dynamics in live cells, complementing genetic knockouts or dominant-negative mutants that often lack temporal control. However, despite their promise, aptamers face hurdles in clinical translation, including in vivo stability and efficient tumor delivery. Future work will focus on improving bioavailability, assessing safety profiles, and exploring combinations with existing therapies to unlock synergistic effects, such as PARP inhibitors, DNA-damaging agents, or immune checkpoint blockade, particularly as persistent DNA damage may stimulate antitumor immunity.
In conclusion, our study provides proof of concept that computationally designed aptamers can disrupt RAD51–BRCA2 interaction—by impairing HR in a dose-dependent manner and inducing SL in BRCA-proficient cancer cells with minimal effects on normal cells—and underscores their potential as powerful tools to exploit DNA repair vulnerabilities in cancer. By sensitizing tumor cells to DNA-damaging agents, aptamers could not only enhance the efficacy of current treatments, but also allow lower chemotherapy doses, reducing side effects while maintaining therapeutic impact. In this scenario, Apt1 exemplifies a first-of-its-kind class of DNA repair inhibitors that complements existing small molecules and peptides, opening new opportunities to expand the reach of SL in oncology.
Methods
Computational design of aptamers
As a starting point for the design of the aptamer we retrieved ChIP-seq peaks for RAD51 available in the ENCODE project under the MCF-7 cancer cell line (https://www.encodeproject.org/experiments/ENCSR442VBJ/). Both Irreproducible Discovery Rate (IDR _thresholded peaks (foreground set) and background peaks used for IDR estimations were utilized. The foreground peaks were filtered for log10(q-value) ≥ 3, while the background peaks were filtered for log10(q-value) ≤ 0.4. From each set, nucleotide fragments of 100 nt and centered around the peak point-source were extracted, resulting in foreground and background sets of 3157 and 67,263 nucleotide fragments, respectively. These two sets were used as input into the DREME algorithm from MEME Suite software (http://meme-suite.org/doc/dreme.html) to identify motifs relatively enriched in the foreground set compared to the background. DREME was configured with a minimum motif length of 4 and a maximum of 15, detecting enriched motifs corresponding to 114 RNA short sequences that were used to create aptamer sequences with a length of approximately 16nt.
For the calculations, two RAD51 regions were defined a (RAD51_Region 1, RAD51_Region 2) as protein fragments with length of ca. 70 amino acids and centered around the interaction pockets between RAD51 and a peptide derived from BRCA2 called BRC485,86. The interaction sites are well defined and the BRC4 structure is well known31,87,88. Region 1 spans from Gly151 to Ile220, and region 2 spans from Asn196 to Ile265. A third RAD51 region of 70 amino acids, spanning from Glu29 to Glu98, was defined as Region 3, centered around the HhH domain (Thr48 to Glu77):
-
RAD51_Region 1 (Supplementary Fig. 1A): GGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTDHQTQLLYQASAMMVESRYALLI
-
RAD51_Region 2 (Supplementary Fig. 1B): NTDHQTQLLYQASAMMVESRYALLIVDSATALYRTDYSGRGELSARQMHLARFLRMLLRLADEFGVAVVI
-
RAD51_Region 3 (Supplementary Fig. 1C): EQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKILAEAAKLVPMGFTTATEFHQRRSE
As a control, we used fragments derived from actin B (Uniprot P60709), a highly expressed cytoskeletal protein in most cell lines. Using a sliding window approach with a 35 amino acid step and a window of 70 amino acids length, a control set of 9 fragments was generated. Then, catRAPID algorithm43,45 was employed to estimate the interaction propensities of the aptamer sequences against Region 1 and Region 2 compared to the control actin B protein fragments (catRAPID score). For each aptamer, mean and standard deviation (SD) of interaction scores against the actin B control protein fragments were estimated. Aptamers were first filtered for catRAPID scores exceeding the minimum threshold that is defined as half a standard deviation above the background mean (threshold=mean_background+0.5*standard_deviation_background; catRAPID_pocket > = threshold). Finally, the filtered aptamers showing a higher catRAPID score against either Region 1 or Region 2 compared to Region 3, were considered as potential aptamers with specificity to RAD51 regions.
Computational prediction of aptamer-protein complexes
The AlphaFold 3 deep-learning framework developed by Google DeepMind and Isomorphic Labs (available as a web server at https://alphafoldserver.com/about) was used to obtain a first in silico validation of the aptamers specificity for RAD51 pockets. This recently updated version of the AlphaFold AI model was chosen since it can generate structure predictions of hetero-complexes starting from one or multiple copies of different types of molecules, including proteins, peptides and nucleic acids, and demonstrated a much higher accuracy for protein-nucleic acid interactions over nucleic acid-specific predictors51.
By means of the AlphaFold Server, several combinations of different molecules and different copy numbers were tested: RAD51 by itself (from Gly21 to Asp339) in 1, 6 and 7 copies; 1 or 6 RAD51 copies with 1, 6, 12 copies of aptamer (i.e. Apt1 and rcApt). For the sake of completeness, the structural prediction of RAD51 oligomeric filament was also run in the presence of both BRC4 (1, 3 or 6 copies) and Apt1 (Supplementary Information).
Each job (producing five different predicted models) was repeated five times on the platform using a different randomly generated seed, for each chosen condition.
Careful visual investigation of the models was performed to verify the quality of the predictions (e.g. absence of steric clashes, loop reconstruction). Given such inspections, the models presented in the Results (as well as in the Supplementary Information) represent the best prediction in terms of predicted template modeling score and interface predicted template modeling score, two of the typical AlphaFold 3 metrics.
For image rendering and contact analysis (reported in Supplementary Table 3), the calculations were run taking advantage of VMD 1.9.4 tools89. Considering the absence of the hydrogens in the models, the threshold distance between the heavy atoms of the protein and the aptamer nucleotides was set at 4 Å in order to include all the putative electrostatic interactions.
Aptamers chemical characteristics
The aptamers here employed (Table 1) were purchased from Eurofins and ATDbio and display lengths varying between 12 to 15 bases. The aptamer called “rcApt” is the reverse complementary sequence of Apt1 and acts as a negative control in these studies. For the BLI experiments, biotin was attached to the 5’ end of all sequences, facilitating molecule binding and detection. For the in-cell assays, locked bases were introduced at both 5’ and 3’ ends of the aptamers Apt1 and rcApt, to increase their stability. In addition, the fluorescent dye Texas Red was conjugated to the 5’ end, providing a visual marker also for FLIM experiments. Prior to each experiment, whether conducted in vitro or in cell, the aptamers were subjected to an incubation period of 1 h at 30 °C to facilitate their correct folding.
Protein production and purification
The full-length human RAD51 protein modified with a 6×His tag at its N-terminus was produced in E. coli Rosetta2(DE3) pLysS cell strain using a pET15b plasmid, containing the gene for ampicillin resistance. The plasmid with the cDNA for 6xHis tag-RAD1 protein was inserted into the Rosetta2(DE3)pLysS by transforming the cells by means of thermal shock treatment, according to literature31. A starter culture was prepared using a single colony of transformed Rosetta2(DE3)pLysS cells, incubated overnight at 37 °C in Luria-Bertani (LB) medium supplemented with 100 µg/mg ampicillin. The starter culture was then scaled up in appropriate volume by diluting it in 100-fold volume of fresh LB, always supplemented with 100 µg/ml ampicillin. During growth, flasks were shaken at 200 rpm and incubated at 37 °C until the cell reached an optical density (OD600) of 0.6 - 0.8. At this point, Isopropyl ß-D-1-thiogalactopyranoside (IPTG) was added to the culture at a final concentration of 0.5 mM to induce heterologous protein production, and the cells were left to grow overnight at 37 °C, while shaking. Cells were collected via centrifugation at 3000 g at 4 °C for 20 min. The pellet was resuspended in a suitable volume (10 mL/gram of wet cell mass) of lysis buffer containing 20 mM Tris-HCl at pH 8.00, 500 mM NaCl, 10 mM imidazole, 5 mM 1,4-Dithiothreitol (DTT) and 10% glycerol by volume, supplemented with a protease inhibitor mix (Protease Inhibitor Cocktail Tablets, EDTA-Free, Sigmafast) and DNAse I (from Merck). Cell lysis was achieved through sonication on ice (14 cycles of 30 s each, and amplitude set at 40%) using a Bandelin Sonoplus HD2070 immersion sonicator. The lysed cell mixture was then centrifuged at 18,000 g at 4 °C for 1 h. Subsequently, the protein extract (supernatant) was introduced into a column packed with nickel-loaded resin, previously equilibrated with buffer A1, which corresponds to lysis buffer without protease inhibitor and DNase I. After the flow-through was discarded, proteins without the His-tag were washed away with a wash buffer (20 mM Tris-HCl, pH 8.00, 500 mM NaCl, 25 mM imidazole, 10% (v/v) glycerol). The target protein was eluted with the same buffer containing 75 mM imidazole. The eluted protein solution was dialyzed overnight at 4 °C in buffer A2 (50 mM Tris-HCl, pH 8.00, 200 mM KCl, 0.25 mM EDTA, 2 mM DTT, 10% (v/v) glycerol) using a dialysis membrane (CarlRoth Cat. N 1991.1) with a cut-off of 10 kDa and then processed through an anion exchange column (ResQ, GE Healthcare) pre-equilibrated with buffer A2. Elution was carried out using a gradient of buffer B2 (50 mM Tris-HCl, pH 8.00, 1 M KCl, 0.25 mM EDTA, 2 mM DTT, 10% (v/v) glycerol), reaching 100% B2 in 10 column volumes. High purity RAD51-containing fractions were pooled and dialyzed against storage buffer (20 mM Hepes, pH 8.00, 250 mM KCl, 0.1 mM EDTA, 2 mM DTT, 10% (v/v) glycerol) overnight at 4°. Protein yield was verified using the optical absorption at 280 nm (extinction coefficient 14,900 M−1 cm−1) of the final sample and was determined to be 200 ng/ml on average. Protein aliquots were flash-frozen and stored at –80 °C.
The BRC4 peptide was synthesized and purchased from Peptide Synthetics Ltd (Fareham, UK). The sequence of the peptide is as follows:
Ac-CKEPTLLGFHTASGKKVKIAKESLDKVKNLFDEKEQ-NH₂
The peptide was N-terminally acetylated and C-terminally amidated to mimic native-like conditions and enhance stability. The purity ( > 95%) was confirmed by analytical HPLC and mass spectrometry, as provided by the manufacturer.
Biolayer interferometry
BLI was employed to determine binding constants (Kds) of the aptamers with RAD51. Experiments were conducted using an Octet Red instrument (ForteBio, Inc., Menlo Park, CA) set at 25 °C. All steps of the assay were executed in binding buffer, a solution of 50 mM Tris HCl buffer at pH 8, containing 200 mM KCl, 0.25 mM EDTA, 1 mM DTT, 10% glycerol and 0.01% Tween-20. Biosensors coated with streptavidin were selected to enable the loading of biotinylated aptamers at a concentration of 2 μg/ml. The protocol to generate the binding curves between the aptamers and RAD51 was set as follows: 180-s baseline; 300-s aptamer loading step on the sensors; 120-s washing step; 600-s association step with increasing protein concentrations (ranging from 20 nM to 20 μM, according to the binding strength); 600-s dissociation step. The experiments were performed with constant shaking at 250 rpm. Kd values were derived by fitting the response intensity (shift in wavelength upon binding) against the protein concentration at a steady state. The experiments were performed in triplicates.
To investigate whether the Apt1 aptamer and the BRC4 peptide bind to the same site on RAD51, a competition assay was performed using BLI. All experiments were conducted in a binding buffer (50 mM Tris-HCl pH 8, 200 mM KCl, 0.25 mM EDTA, 1 mM DTT, 10% glycerol, and 0.01% Tween-20). Streptavidin-coated biosensors were used for immobilization of biotinylated Apt1 at a concentration of 2 μg/ml. RAD51 (1 μM) was incubated with increasing concentrations of BRC4 peptide (0–20 μM) to assess competition. The assay protocol included the following steps: 180 s baseline equilibration, 200 s aptamer loading, 120 s washing, 400 s association with RAD51/BRC4, and 400 s dissociation. All measurements were performed at 25 °C with continuous shaking at 250 rpm.
Negative staining transmission electron microscopy
Recombinant RAD51 protein (2.5 μM) was incubated at 4 °C for 1 h, either by itself or in the presence of increasing concentrations of Apt1. After incubation, each sample (5 µL) was adsorbed onto pure carbon film 300-mesh copper grids (Electron Microscopy Sciences, Hatfield, PA, USA). Following several washes in the glycerol-free storage buffer (20 mM Hepes, pH 8.00, 250 mM KCl, 0.1 mM EDTA, 2 mM DTT), each sample was negatively stained using 1% uranyl acetate in Milli-Q water. The samples were observed using a JEM-1011 (JEOL) transmission electron microscope (TEM) equipped with a thermionic source (W filament) and a maximum acceleration voltage of 100 kV. The microscope was fitted with a Gatan Orius SC1000 series CCD camera (4008 × 2672 active pixels), fiber optically coupled to a high-resolution phosphor scintillator. The average diameter of the ring-like structures, identified in the RAD51:Apt1 = 1:6 condition, was calculated using the tool analyze and measure of the image analysis software ImageJ.
Fluorescence-lifetime imaging microscopy
FLIM images have been acquired using a Digital Frequency Domain (DFD) FLIMbox (ISS inc., Champaign, IL) coupled to a A1RMP multiphoton microscope (Nikon, Japan), calibrated with a 10 µM solution of Coumarin 153 in methanol (τ= 4.3 ns). Samples were 2-photon excited focusing a Chameleon UltraII Ti:Sapphire laser (Coherent inc., Saxonburg, PA) tuned at 850 nm through a 60x oil objective (NA = 1.45). Fluorescence emitted photons were filtered using a 525/50 nm BP filter for Coumarin 153 and a 605/70 nm for Texas Red labeled aptamers. The analyses were performed with ISS VistaVision multi-image phasor analysis software, where sine (s) and cosine (g) coordinates are defined as
where m is the modulation and φ the phase acquired of the h-th pixel in the FLIM image and F the laser repetition rate.
Cell culture handling
BxPC-3, HEK293 and MCF-10A cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). BxPC-3 cells were grown in RPMI 1640, supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, and 10% fetal bovine serum (FBS). HEK293 cells were grown in DMEM high-glucose, supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, and 10% FBS. MCF-10A were grown in DMEM/F-12 (1:1), supplemented with 2.5 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, 10 μg/mL human insulin (#12585, Gibco), 10 ng/mL epidermal growth factor (EGF) (#E9644, Sigma-Aldrich), 100 ng/mL cholera toxin (#C8052, Sigma-Aldrich), 0.5 μg/mL hydrocortisone (#H0888, Sigma-Aldrich) and 5% horse serum (#16050-130, Gibco). Cells were maintained at 37 °C in a humidified 5% CO2 atmosphere, split when at confluence and routinely tested for Mycoplasma contamination. All culture media and supplements, unless otherwise specified, were obtained from Sigma Aldrich (St. Louis, MO, USA). Olaparib (PubChem CID: 23725625, #S1060) and cis-diamminedichloroplatinum(II) (cisplatin) (PubChem CID: 2767, #S1166) were purchased from Selleck Chemicals (Houston, TX, USA). Olaparib was dissolved in DMSO at the final concentration of 10 mM. Cisplatin was dissolved in 1X PBS at the final concentration of 5 mM. Stock aliquots were stored at −20 °C and diluted in complete medium prior to each experiment.
IMMUNOFLUORESCENCE SAMPLE PREPARATION FOR RAD51 BEHAVIOR INVESTIGATION
For microscopy analysis, cells were seeded at a concentration of 150,000 cells/mL onto 24-well plates with coverslips. After 24 h, complete medium was replaced with FBS-free medium for 18 h-24 h for cell synchronization. Subsequently, cells were recovered for 2 h in fresh complete medium and transfected with 0.5 μg/ml aptamers using Lipofectamine® 2000 (#11668019, Thermo Fisher Scientific) following the manufacturer’s instructions. After transfection, cells were allowed to recover overnight before further treatments. The day after transfection, cells were treated with 50 µM cisplatin for 2 h to induce DNA damage. Following cisplatin treatment, cells were rinsed twice with 1X phosphate-buffered saline (PBS) and then fixed for 10 min at room temperature using a 4% paraformaldehyde (PFA) solution in PBS. Cells were then washed three times with 1X PBS permeabilized with 0.1% Triton-X 100 in 1X PBS for 5 min, and washed three times in 1X PBS.
Cells were incubated with 5% bovine serum albumin (BSA) in PBS for 30 min at room temperature to block nonspecific binding sites. Following the blocking step, cells were incubated with a rabbit anti-RAD51 primary antibody (Bio Academia, 70-001), diluted 1:1000 in 5% BSA/PBS, for 1 h at room temperature. After washing, cells were incubated with a mouse anti-BRCA2 primary antibody (Santa Cruz Biotechnology, sc-518154), diluted 1:250 in 5% BSA/PBS, for 1 h at room temperature. Subsequently, an Alexa Fluor® 488-conjugated goat anti-rabbit secondary antibody (Thermo Fisher, A-11008) was applied at a 1:1000 dilution in 5% BSA/PBS and incubated for 1 h at room temperature in the dark. This was followed by incubation with an Alexa Fluor® 594-conjugated goat anti-mouse secondary antibody (Thermo Fisher, A11032), also diluted 1:1000 in 5% BSA/PBS, for 1 h at room temperature in the dark.
After three washes with PBS, cells were treated for 5 min with a 0.5 μg/ml solution of 4′,6-diamidino-2-phenylindole (DAPI) prepared in PBS. Following three additional rinses in PBS, the coverslips were positioned face-down on glass slides using ProLong™ Diamond Antifade Mountant (Thermofisher, P36965).
Immunofluorescence sample preparation for DNA damage quantification
To evaluate the effect of aptamers on physiological DSBs, cells were analyzed three days after transfection by assessing those positive for γH2AX foci. To determine the aptamer impact on SL, olaparib was added at a concentration of 10 µM one day post aptamer transfection. The formation γH2AX foci was analyzed three days after transfection. In both cases, cells were rinsed two times with PBS and then fixed for 10 min at room temperature using a 4% paraformaldehyde solution prepared in PBS. Subsequent to three PBS washes, cells underwent permeabilization with 0.1% Triton-X 100 in PBS for a duration of 5 min, followed by three more PBS washes. Cells were treated with 5% BSA prepared in PBS for 30 min as a blocking step. Following this, a primary antibody against phospho-histone H2AX (Ser139), clone JBW301 (Merck/Sigma Aldrich, 05-636), was employed at a 1:1000 dilution in 5% BSA/PBS and incubated for 1 h at room temperature. Subsequently, cells were treated with a secondary anti-mouse antibody conjugated to Alexa Fluor® 647 (Thermofisher, A-21236) at a 1:1000 dilution and incubated for an additional hour at room temperature.
After three washes with PBS, cells were treated for 5 min with a 0.5 μg/ml solution of 4′,6-diamidino-2-phenylindole (DAPI) prepared in PBS. Following three additional rinses in PBS, the coverslips were positioned face-down on glass slides using ProLongTM Diamond Antifade Mountant (Thermofisher, P36965).
Immunofluorescence images acquisition
To analyze RAD51 and BRCA2 fluorescence, slides containing fixed cells were examined using Nikon’s NSPARC confocal microscope equipped with a Plan Apo Lambda S 40x/1.25 NA silicon immersion objective. Images were acquired with a projection zoom of 1 Airy Unit (AU) pixel size 70 nm and dwell time 0.4 µsec, DAPI was excited using a 405 nm laser with 4% power transmission, fluorescence collected in the range 430 - 463 nm, Alexa Fluor®488 was excited using a 488 nm with 4% power transmission, fluorescence collected in the range 503 - 545 nm, Alexa Fluor® 563 was excited using a 561 nm with 10% power transmission, fluorescence collected in the range 582 - 618 nm. For each experimental condition, a minimum of eight representative images were collected. Quantitative analysis of fluorescence intensity and foci formation was performed on at least 200 nuclei per sample.
For γH2AX detection, images were acquired using Nikon’s A1R confocal microscope with a Plan Apo TIRF 60x/1.49 NA oil immersion objective (DIC H N2). Images were acquired at pinhole size of 1 AU, pixel size of 104 nm and scan speed 0.125, excitation of DAPI was performed at 405 nm with 10% laser power transmission, fluorescence collected in the range 430-475 nm, Alexa Fluor® 488 was excited at 488 nm and laser power set at 2.5% of transmission, fluorescence collected in the range 500-550 nm, Alexa Fluor® 563 was excited at 561 nm and laser power set at 3% of transmission, fluorescence collected in the range 570-615 nm. For each condition, a minimum of eight representative images were analyzed. Quantification of γH2AX foci formation was performed on at least 500 nuclei per sample.
Immunofluorescence images analysis
Our in-house macro was run on the software ImageJ to perform fluorescence intensity analysis on images of fluorophores associated with RAD51 and BRCA2, with and without cisplatin and in the absence/presence of fluorophore-tagged locked nucleic acid (LNA) Apt1 or rcApt, to assess the impact of aptamers on the DSB repair mechanism. To measure the intensity of nuclear RAD51 fluorescence, the nuclear region was identified using the nuclear stain DAPI. To segment the nuclei, a Gaussian filter was applied to the images to smooth them and reduce noise. The image threshold was adjusted (Otsu method) to differentiate between the signal of interest and background. Finally, a watershed algorithm was applied to the binary image to separate nuclei that might be touching each other. Particle analysis parameters were set as follows: size range 1000-Infinity pixels; circularity between 0.4 and 1. Once identified in the nuclear region and added to the region of interest (ROI) manager, nuclear fluorescence was quantified by mean intensity values in the green and red channels, corresponding to RAD51 and BRCA2. The colocalization analysis by measuring the Manders coefficient was performed by ImageJ and the tool JACoP90. For each condition, one image was selected and processed with ImageJ’s algorithms “Smooth” and “Subtract Background”, rolling bar set at 50 pixels and all other features disabled. In each image, 10 nuclei were randomly selected and analysis performed with an intensity threshold selected by measuring the mean intensity value of the two channels in the untreated images; the values were set at 20 for the green channel (RAD51) and 10 for the red channel (BRCA2). The fluorescence data were normalized by nuclear area. To estimate the fluorescence intensity ratio between the cytoplasmic and nuclear RAD51 signal, a macro was designed for ImageJ with the following operational pipeline. The program was configured to measure the area and the mode of fluorescence intensity of ROIs representing nuclear and cytoplasmic areas in the images. A Gaussian blur with Sigma 1 was applied to the DAPI channel, aiding in the segmentation of the nuclear region with higher confidence. The image was segmented using the Otsu criterion, an automatic and robust threshold parameter. Holes in the binary images were filled using the ImageJ algorithm, and these were used to create a region of interest. The second channel -the signal from RAD51- was processed in a similar manner. A Gaussian blur with sigma 0.5 was applied, and the image was segmented using the MinError function; this allowed for the obtaining of a binary image of the entire cell due to the RAD51 distribution. The binary image of the nucleus was subtracted from that of the whole cells to obtain the cytoplasmic area image. Finally, the mode intensity and area of the two regions of interest, the cytoplasmic and nuclear, were measured. The ratio between the nuclear and cytoplasmic signal was used to discriminate between control and treated cells as previously described. The fluorescence values of nuclear RAD51 are calculated by dividing the nuclear mean fluorescence by the nuclear area and are reported as percentage. Mean/area of cells treated with cisplatin only is considered to be 100%. To assess the increased presence of γH2AX foci within nuclei due to DNA damage, a macro was developed for ImageJ, establishing a comprehensive operational protocol for automated image analysis across a specified directory. Image parameters, such as the directory path, file format, and channels designated for segmentation and analysis, were configured at the outset. A reference image, specifically one of cells treated with cisplatin, was selected to adjust threshold parameters essential for segmenting the signal of interest. Subsequent to this initial setup, a Gaussian blur with a sigma value of 1 was applied to the DAPI channel to enhance the segmentation accuracy of the nuclear regions. The segmentation was executed employing the Otsu criterion, an automated and robust thresholding method. Holes within the binary images were filled using ImageJ’s algorithm, facilitating the delineation of individual nuclei as regions of interest (ROIs) through the Analyze Particles function. Parameters were set to include objects with a size of at least 5000 pixels, a circularity range of 0.5–1.00, and to exclude objects at the image periphery.For the analysis of the γH2AX signal, a Gaussian blur with a sigma of 0.5 was applied, followed by segmentation using threshold values determined from the reference image. This process generated a binary image of the γH2AX foci. Each nuclear ROI containing foci was quantified by calculating the mean pixel value within the binary γH2AX image; a mean exceeding one was indicative of a positive ROI. The proportion of positive nuclei to the total number of nuclei served to distinguish between control and treated cells, as delineated in the study. A custom ImageJ macro was developed to quantify the number of micronuclei. Two distinct particle populations were segmented based on user-specified size and circularity parameters: (i) small micronuclei, and (ii) nuclei. Image preprocessing steps included contrast enhancement, background subtraction, thresholding using the Otsu method, hole filling, and watershed separation to refine object boundaries. Particles and nuclei were then identified with the Analyze Particles function, and their respective counts were stored. For each image, the macro calculated the ratio of micronuclei to nuclei, providing a normalized measure of nuclear micronuclei frequency.
Cell viability assessment
2D cell lines
Cell viability in 2D cell cultures was assessed via MTT [3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay as described previously91. Each transfection was carried out using Lipofectamine® 2000 following the manufacturer’s instruction. BxPC-3 (75,000 cells/mL) and MCF-10A (50,000 cells/mL) cells were seeded at a volume of 100 μL/well in 96-well plates and allowed to adhere overnight. After 24 h, cells were transfected with Apt1 or rcApt (50, 100 and 200 nM, corresponding to 0.25, 0.5 and 1 μg/mL) alone or concurrently treated with 10 μM olaparib. After 72 h, a sterile solution of 5 mg/mL MTT in 1X PBS was added to each well at a final concentration of 0.5 mg/mL. Plates were incubated at 37 °C for 4 h, and the formazan crystals were solubilised overnight by adding a 1:1 volume of 10% SDS/0.01 M HCl solution to each well. Absorbance was measured on a Tecan Spark® multiplate reader at 570 nm and 690 nm wavelengths.
3D spheroids
MTT assay was used to assess cell viability also in 3D spheroids as previously employed57. BxPC-3 cells (300,000 cells/mL) were seeded at a volume of 100 μL/well in 96-multiwell plates coated with sterile 1% agarose dissolved in 1X PBS, grown for 96 h and treated for 72 h as indicated for the 2D cell cultures. After treatment, cell viability was assessed following the same protocol detailed above.
Drug interaction index calculation
The potential synergistic effect of aptamer and olaparib association in the employed cell lines was evaluated by calculating the drug interaction index (i. index) according to the Fischel et al. adaptation of the Chou and Talalay method as previously reported57:
where: i. index <0.8: synergism; 0.8 <i. index <1.2: additive effect; i. index > 1.2: antagonism.
Cell death assessment with vital dyes
Live/dead staining was performed as previously described57. For cell death assessment with vital dyes in 3D cell cultures, BxPC-3 spheroids were obtained and treated with the aptamers, alone or in combination with 10 μM olaparib, as detailed in the “Cell viability assessment” section. After treatment, calcein acetoxymethyl ester (Calcein-AM) (#C3099, Thermo Fisher Scientific) and propidium iodide (PI) (#P4846, Sigma-Aldrich) were added to each well at the final concentration of 2.5 and 3.75 μM, respectively. To determine cell death rate, Calcein-AM (Ex = 485 nm, Em = 530 nm) and PI (Ex = 535 nm, Em = 620 nm) fluorescence signals were measured on a Tecan Spark® multiplate reader and results were expressed as PI/Calcein-AM ratio.
Homologous recombination efficiency evaluation
Homologous recombination quick assay (HR-QA)
HR was assessed with the commercially available Homologous Recombination Assay Kit (#35600, Norgen, Thorold, ON, Canada) as previously reported57. Each transfection was carried out using Lipofectamine® 2000 following the manufacturer’s instruction. BxPC-3 cells (400,000 cells/mL) were seeded at a volume of 500 μL/well in a 24-multiwell plate, allowed to adhere overnight and then transfected with Apt1 or rcApt 100 nM (i.e. 0.5 μg/mL). After 24 h, the two assay plasmids (dl-1 and d-2) were co-transfected following the manufacturer’s instructions for 5 h, washed with 1X PBS, harvested, and DNA was isolated using Illustra Tissue and Cell Genomic Prep Mini Spin kit (GE Healthcare). Sample concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). HR efficiency was assessed via real-time PCR, performed on a ViiA™ 7 Real-Time PCR System (Applied Biosystems), using iTaq™ Universal SYBR® Green Supermix (#1725124, Bio-Rad Laboratories, Inc.), 25 ng of template, the primer mixtures included in the assay kit and following the protocol indicated by the manufacturer. Data analysis was based on the 2-ΔΔCt method92: [recombination product/backbone plasmids]treated versus [recombination product/backbone plasmids]control.
mClover-Lamin A-based Homologous Recombination assay (mCL-HR)
The mCL-HR assay was performed as previously detailed57. Each transfection was carried out using Lipofectamine® 2000 following the manufacturer’s instruction. HEK293 cells (60,000 cells/mL) were seeded at a volume of 1 mL/well in a 12-multiwell plate with glass coverslips, allowed to adhere overnight and then transfected with Apt1 or rcApt 50, 100 and 200 nM (i.e. 0.25, 0.5 and 1 μg/mL). After 24 h, cells were transfected with 500 ng sgRNA plasmid targeting Lamin A (pUC CBA-SpCas9.EF1a-BFP.sgLMNA, #98971, Addgene) and 500 ng donor plasmid (pCAGGS Donor mClover-LMNA, #98970, Addgene). After 72 h, cells were fixed with a 4% paraformaldehyde (PFA) solution in 1X PBS for 15 min, stained with 1 μg/mL Hoechst 33342 (#62249, Thermo Fisher Scientific) and washed twice before mounting with Dako Faramount Aqueous Mounting Medium (#S3025, Agilent Technologies, Santa Clara, CA, US). Fluorescence images were taken using a Leica K7 Color CMOS Microscope Camera through a Leica DM5500 B microscope (Leica Microsystems, Wetzlar, Germany) equipped with filters Y5, L5 and DAPI. Images were analyzed with ImageJ software (W. Rasband, Research Service Branch, National Institute of Mental Health, National Institutes of Health, Bethesda, MD and Laboratory for Optical and Computational Instrumentation, University of Wisconsin) to obtain mClover-positive nuclei/total nuclei ratios for each condition, then expressed as percentage relative to control cells.
Spheroid fluorescence and brightfield imaging
Fluorescence images of live/dead stained 3D spheroids were taken using a Leica DFC360 FX Camera through a Leica DMI6000 B inverted microscope (Leica Microsystems, Wetzlar, Germany) equipped with filters for FITC, TRITC and DAPI (scale bars were added using the ImageJ software). Brightfield imaging of 3D spheroids was performed at 0, 24, 48 and 72 h treatment timings to determine spheroid volume change. Pictures were taken using a Leica DFC360 FX camera through a Leica DMI6000 B inverted microscope (scale bars were added using the ImageJ software). Image processing was performed with ImageJ software and spheroid volume was calculated using the following general formula (as previously described57,: Spheroid Volume (V) = 0.5 ∙(Length)∙(Width)2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are in Source data file and the immunofluorescence image have been deposited in the Zenodo database, publicly accessible under the following accession codes: https://doi.org/10.5281/zenodo.17413480 - Immunofluorescence images showing the effect of increasing concentrations of Apt1 on RAD51 and BRCA2 nuclear localization in response to DNA damage (Fig. 5A). https://doi.org/10.5281/zenodo.17401800 – Immunofluorescence images showing the percentage of BxPC-3 cells positive for γH2AX foci 72 h after transfection with Apt1 or rcApt, alone or in combination with 10 μM olaparib (Fig.6C). https://doi.org/10.5281/zenodo.17398501 – Immunofluorescence images collected to investigate the effect of Apt1 on RAD51 nuclear localization in response to DNA damage (Supplementary Fig. 11). Source data are provided with this paper.
References
HuangZhou, P. K. & Zhou, P.-K. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target Ther. 6, 254 (2021).
AlhmoudMalki, M. I., Woolley, J. F., Al Moustafa, A.-E. & Malki, M. I. DNA damage/repair management in cancers. Cancers 12, 1050 (2020).
HanahanWeinberg, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
YaoDai, Y. & Dai, W. Genomic Instability and Cancer. J Carcinog Mutagen 5 (2014).
Chen, J. et al. Exploring DNA damage and repair mechanisms: a review with computational insights. BioTech. (Basel) 13, 3 (2024).
ScullyWillis, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
ThadathilKhan, N., Hori, R., Xiao, J. & Khan, M. M. DNA double-strand breaks: a potential therapeutic target for neurodegenerative diseases. Chromosome Res. 27, 345–364 (2019).
San FilippoKlein, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008).
CarverZhang, A. & Zhang, X. Rad51 filament dynamics and its antagonistic modulators. Semin. Cell Dev. Biol. 113, 3–13 (2021).
BonillaBernstein, B., Hengel, S. R., Grundy, M. K. & Bernstein, K. A. RAD51 gene family structure and function. Annu. Rev. Genet. 54, 25–46 (2020).
ReymerNordén, A., Frykholm, K., Morimatsu, K., Takahashi, M. & Nordén, B. Structure of human Rad51 protein filament from molecular modeling and site-specific linear dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 106, 13248–13253 (2009).
Jensen, R. B., Carreira, A. & Kowalczykowski, S. C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 (2010).
LiuHeyer, J., Doty, T., Gibson, B. & Heyer, W.-D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 (2010).
Thorslund, T. et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 (2010).
Kowalczykowski, S. C. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410 (2015).
Sánchez, H. et al. Architectural plasticity of human BRCA2-RAD51 complexes in DNA break repair. Nucleic Acids Res. 45, 4507–4518 (2017).
TrennerSartori, A., Godau, J. & Sartori, A. A. A short BRCA2-derived cell-penetrating peptide targets RAD51 function and confers hypersensitivity toward PARP inhibition. Mol. Cancer Ther. 17, 1392–1404 (2018).
Bagnolini, G. et al. Identification of RAD51-BRCA2 inhibitors using N-acylhydrazone-based dynamic combinatorial chemistry. ACS Med. Chem. Lett. 13, 1262–1269 (2022).
Pellegrini, L. et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420, 287–293 (2002).
LordAshworth, C. J. & Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 16, 110–120 (2016).
Bagnolini, G. et al. Synthetic lethality in pancreatic cancer: Discovery of a new RAD51-BRCA2 small molecule disruptor that inhibits homologous recombination and synergizes with olaparib. J. Med. Chem. 63, 2588–2619 (2020).
Ryan, C. J., Devakumar, L. P. S., Pettitt, S. J. & Lord, C. J. Complex synthetic lethality in cancer. Nat. Genet. 55, 2039–2048 (2023).
Nijman, S. M. B. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett. 585, 1–6 (2011).
Deeks, E. D. Olaparib: first global approval. Drugs 75, 231–240 (2015).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Fu, X. et al. Mechanism of PARP inhibitor resistance and potential overcoming strategies. Genes Dis. 11, 306–320 (2024).
RoseBolderson, M., Burgess, J. T., O’Byrne, K., Richard, D. J. & Bolderson, E. PARP inhibitors: Clinical relevance, mechanisms of action and tumor resistance. Front. Cell Dev. Biol. 8 (2020).
Falchi, F. et al. Synthetic lethality triggered by combining Olaparib with BRCA2-Rad51 disruptors. ACS Chem. Biol. 12, 2491–2497 (2017).
ElyashivLedermann, O., Wong, Y. N. S. & Ledermann, J. A. Frontline Maintenance Treatment for Ovarian Cancer. Curr. Oncol. Rep. 23, 97 (2021).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
Schipani, F. et al. The mechanistic understanding of RAD51 defibrillation: a critical step in BRCA2-mediated DNA repair by homologous recombination. Int. J. Mol. Sci. 23, 8338 (2022).
Previtali, V. et al. New Horizons of synthetic lethality in cancer: current development and future perspectives. J. Med. Chem. 67, 11488–11521 (2024).
Scott, D. E. et al. A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death. Cell Chem. Biol. 28, 835–847.e5 (2021).
EllingtonSzostak, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
TuerkGold, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Gao, F. et al. Recent advances in aptamer-based targeted drug delivery systems for cancer therapy. Front Bioeng. Biotechnol. 10, 972933 (2022).
KeefeEllington, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 9, 537–550 (2010).
DomsicovaBreier, M., Korcekova, J., Poturnayova, A. & Breier, A. New insights into aptamers: an alternative to antibodies in the detection of molecular biomarkers. Int. J. Mol. Sci. 25, 6833 (2024).
ZhouRossi, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 (2017).
Zacco, E. et al. Probing TDP-43 condensation using an in silico designed aptamer. Nat. Commun. 13, 3306 (2022).
DastiTartaglia, A., Cid-Samper, F., Bechara, E. & Tartaglia, G. G. RNA-centric approaches to study RNA-protein interactions in vitro and in silico. Methods 178, 11–18 (2020).
Spence, H. et al. RNA aptamer reveals nuclear TDP-43 pathology is an early aggregation event that coincides with STMN-2 cryptic splicing and precedes clinical manifestation in ALS. Acta Neuropathol. 147, 50 (2024).
ArmaosTartaglia, A., Colantoni, A., Proietti, G., Rupert, J. & Tartaglia, G. G. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 49, W72–W79 (2021).
Luige, J. et al. Design and characterization of G-quadruplex RNA aptamers reveal RNA-binding by KDM5 lysine demethylases. Comput. Struct. Biotechnol. J. 27, 2719–2729 (2025).
BellucciTartaglia, M., Agostini, F., Masin, M. & Tartaglia, G. G. Predicting protein associations with long noncoding RNAs. Nat. Methods 8, 444–445 (2011).
Cirillo, D. et al. Quantitative predictions of protein interactions with long noncoding RNAs. Nat. Methods 14, 5–6 (2016).
Martinez, S. F. et al. Targeting human Rad51 by specific DNA aptamers induces inhibition of homologous recombination. Biochimie 92, 1832–1838 (2010).
Agostini, F. et al. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics 29, 2928–2930 (2013).
Cerase, A. et al. Phase separation drives X-chromosome inactivation: a hypothesis. Nat. Struct. Mol. Biol. 26, 331–334 (2019).
Sanchez de Groot, N. et al. RNA structure drives interaction with proteins. Nat. Commun. 10, 3246 (2019).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Brouwer, I. et al. Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA. EMBO J. 37 (2018).
Short, J. M. et al. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res. 44, gkw783 (2016).
ApplebyPellegrini, R., Joudeh, L., Cobbett, K. & Pellegrini, L. Structural basis for stabilisation of the RAD51 nucleoprotein filament by BRCA2. Nat. Commun. 14, 7003 (2023).
De Los Santos, C., Chang, C. W., Mycek, M. A.-W. & Cardullo, R. A. Detection and analysis of cellular dynamics on a molecular scale using fluorescence microscopy. Mol. Reprod. Dev. 82, 587–604 (2015).
Becker, W. Fluorescence lifetime imaging-techniques and applications. J. Microsc. 247, 119–136 (2012).
Masi, M. et al. Investigating synthetic lethality and PARP inhibitor resistance in pancreatic cancer through enantiomer differential activity. Cell Death Discov. 11, 106 (2025).
Stewart, M. D. et al. Homologous recombination deficiency: Concepts, definitions, and assays. Oncologist 27, 167–174 (2022).
MladenovTsaneva, E., Anachkova, B. & Tsaneva, I. Sub-nuclear localization of Rad51 in response to DNA damage. Genes Cells 11, 513–524 (2006).
Brinkman, E. K. et al. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol. Cell 70, 801–813.e6 (2018).
Yamashita, N. et al. Cumulative DNA damage by repeated low-dose cisplatin injection promotes the transition of acute to chronic kidney injury in mice. Sci. Rep. 11, 20920 (2021).
Rocha, C. R. R., Silva, M. M., Quinet, A., Cabral-Neto, J. B. & Menck, C. F. M. DNA repair pathways and cisplatin resistance: an intimate relationship. Clinics 73, e478s (2018).
Castello, M. et al. A robust and versatile platform for image scanning microscopy enabling super-resolution FLIM. Nat. Methods 16, 175–178 (2019).
Sun, H. et al. CRIP1 cooperates with BRCA2 to drive the nuclear enrichment of RAD51 and to facilitate homologous repair upon DNA damage induced by chemotherapy. Oncogene 40, 5342–5355 (2021).
Roberti, M. et al. Rad51/BRCA2 disruptors inhibit homologous recombination and synergize with olaparib in pancreatic cancer cells. Eur. J. Med. Chem. 165, 80–92 (2019).
Myers, S. H. et al. An 19F NMR fragment-based approach for the discovery and development of BRCA2-RAD51 inhibitors to pursuit synthetic lethality in combination with PARP inhibition in pancreatic cancer. Eur. J. Med. Chem. 265, 116114 (2024).
Tan, M. H. et al. Characterization of a new primary human pancreatic tumor line. Cancer Invest 4, 15–23 (1986).
Cowley, G. S. et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data 1, 140035 (2014).
CrowleyO’Reilly, F., Park, W. & O’Reilly, E. M. Targeting DNA damage repair pathways in pancreas cancer. Cancer Metastasis Rev. 40, 891–908 (2021).
RayRaghavan, U. & Raghavan, S. C. Understanding the DNA double-strand break repair and its therapeutic implications. DNA Repair (Amst.) 106, 103177 (2021).
KhannaJackson, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 (2001).
Patel, J. et al. DNA damage and mitochondria in cancer and aging. Carcinogenesis 41, 1625–1634 (2020).
SchumacherHoeijmakers, B., Pothof, J., Vijg, J. & Hoeijmakers, J. H. J. The central role of DNA damage in the ageing process. Nature 592, 695–703 (2021).
Sharma, A., Singh, K. & Almasan, A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 920, 613–626 (2012).
SachdevMita, E., Tabatabai, R., Roy, V., Rimel, B. J. & Mita, M. M. PARP inhibition in cancer: an update on clinical development. Target. Oncol. 14, 657–679 (2019).
O’Connor, M. J. Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560 (2015).
Longati, P. et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 13, 95 (2013).
Kota, S. et al. A novel three-dimensional high-throughput screening approach identifies inducers of a mutant KRAS selective lethal phenotype. Oncogene 37, 4372–4384 (2018).
Lai, E. et al. BRCA-mutant pancreatic ductal adenocarcinoma. Br. J. Cancer 125, 1321–1332 (2021).
Klein, H. L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair 7, 686–693 (2008).
ZhangRen, X., Ma, N., Yao, W., Li, S. & Ren, Z. RAD51 is a potential marker for prognosis and regulates cell proliferation in pancreatic cancer. Cancer Cell Int. 19, 356 (2019).
HenningStürzbecher, W. & Stürzbecher, H. W. Homologous recombination and cell cycle checkpoints: Rad51 in tumour progression and therapy resistance. Toxicology 193, 91–109 (2003).
ZeviniHiscott, A., Olagnier, D. & Hiscott, J. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 38, 194–205 (2017).
ShraimHunaiti, A. S., Abdel Majeed, B. A., Al-Binni, M. A. & Hunaiti, A. Therapeutic potential of aptamer-protein interactions. ACS Pharmacol. Transl. Sci. 5, 1211–1227 (2022).
ScottHyvönen, D. E., Marsh, M., Blundell, T. L., Abell, C. & Hyvönen, M. Structure-activity relationship of the peptide binding-motif mediating the BRCA2:RAD51 protein-protein interaction. FEBS Lett. 590, 1094–1102 (2016).
LoBlundell, T., Pellegrini, L., Venkitaraman, A. R. & Blundell, T. L. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair 2, 1015–1028 (2003).
Miron, S. et al. DMC1 and RAD51 bind FxxA and FxPP motifs of BRCA2 via two separate interfaces. Nucleic Acids Res. 52, 7337–7353 (2024).
RajendraVenkitaraman, E. & Venkitaraman, A. R. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 38, 82–96 (2010).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–8, 27–8 (1996).
Bolte Cordelières, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Buoso, E. et al. Disruption of epithelial barrier integrity via altered GILZ/c-Rel/RACK1 signaling in inflammatory bowel disease. J. Crohns. Colitis 19 (2025).
Masi, M. et al. The labyrinthine landscape of APP processing: State of the art and possible novel soluble APP-related molecular players in Traumatic Brain Injury and neurodegeneration. Int. J. Mol. Sci. 24, 6639 (2023).
Acknowledgements
The authors would like to thank the other members of Tartaglia and Cavalli’s groups. The research leading to this work was supported by the ERC ASTRA_855923 (G.G.T.), EIC Pathfinder IVBM4PAP_101098989 (G.G.T.) and PNRR grant from the National Centre for Gene Therapy and Drugs based on RNA Technology (CN00000041 EPNRRCN3 (G.G.T. and A.C.). F.D.P. acknowledges the support granted by the European Union—Next Generation EU, Mission 4 Component 1 CUP D53D23016360001, PRIN-PNRR, Grant n. P2022CLXMK, for his current position. M.M. is supported by an AIRC Foundation for Cancer Research NGO Fellowship−Italy Post-doc 2022 (Project Code: 28174). L.B. is supported by a Marie Skłodowska-Curie post-doctoral fellowship (UNDERPIN_101063903). The authors extend their gratitude to Giuseppe Vicidomini and Eleonora Perego for their help with the microscopy experiments. The authors also acknowledge the fundamental support of the Nikon Imaging Centre and the Electron Microscopy facility at Fondazione Istituto Italiano di Tecnologia. Special thanks are due to Prof. Marco De Vivo and Francesca Pellegrino for producing the PolH protein, which was tested in our internal controls.
Author information
Authors and Affiliations
Contributions
G.M. performed the experiments, participated in their design and wrote the manuscript. E.Z. performed and supervised the experiments and wrote the manuscript. M.M. designed and supervised the cell experiments, carried out the SL experiments and wrote the manuscript. A.A. and G.G.T. generated the aptamers, G.G.T. F.D.P. carried out the structural predictions and analysis of RAD51 complexed with aptamers, and wrote the manuscript. M.G. and L.B. provided assistance with cellular experiments. J.R. contributed to in vitro experimental work. G.V. was involved in producing some of the recombinant protein. M.O. developed the macro used for in-cell analysis. M.S. worked on the FLIM experiment. R.M. worked on the T.E.M. experiment. S.G. designed and supervised the experiments and wrote the manuscript. AC formulated the initial hypothesis and supervised the work. G.G.T. formulated the initial hypothesis, supervised the work and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A patent on the aptamers has been filed (PCT/IB2025/059812, Italian priority application N. IT102024000004522) on which G.G.T., A.C., E.Z., G.M., A.A., M.G., J.R., S.G., M.O., M.S., and M.M. are named as inventors. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Vittorio de Franciscis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Milordini, G., Zacco, E., Masi, M. et al. Computationally-designed aptamers targeting RAD51-BRCA2 interaction impair homologous recombination and induce synthetic lethality. Nat Commun 17, 70 (2026). https://doi.org/10.1038/s41467-025-66694-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66694-9