Abstract
Current colorectal cancer mouse models either lack colon specificity, limiting progression towards more advanced disease, or preclude evaluation of resident stem cells as cancer origins. Here we report the identification of NOX1 and NPY1R as cell-surface markers enriched in LGR5+ stem cells predominantly within the caecum and exclusively within the middle and distal colorectum, respectively. Selective dysregulation of Wnt signalling in NOX1+ or NPY1R+ stem cells using CreERT2 mouse lines drives colon cancer initiation, predominantly within the caecum and rectum respectively, establishing these stem cell populations as important sources of colon cancer. Selective conditional activation of Wnt signalling and oncogenic Kras in combination with loss of TRP53 in these stem cell compartments resulted in the development of advanced, invasive cancers. This study establishes CreERT2 drivers as valuable tools for studying stem cell contributions to colon cancer.
Similar content being viewed by others
Main
The epithelial lining of the gastrointestinal tract is essential for completing food digestion and mediating the efficient absorption of nutrients while also providing critical barrier functions and innate immune defence against potentially harmful microbes. Its extremely harsh operating environment imposes an obligate requirement for constant renewal, driven by discrete populations of stem cells residing within specialized niches at the base of deep invaginations within the epithelium known as crypts1. The functional units of the small intestine comprise many millions of finger-like villi, each associated with several crypts that collectively supply the new functional epithelial lineages needed to replace the billions of cells being extruded at villus tips daily. In contrast, the lining of the more distal part of the gastrointestinal tract, the colon, comprises multiple crypts that supply new functional lineages to a contiguous layer of flat surface epithelium. The colon is organized into anatomically distinct regions comprising the caecum, which directly connects the small intestine, followed by proximal, middle and distal compartments that terminate in the rectum and anus. Subtle differences in the architecture, composition and renewal rates of the epithelia from different regions of the colon, together with marked variations in their resident microbiome reflect their functional specialization2,3,4. Although the functions of the different compartments of the colon are not fully understood, the caecum is thought to play a role in water and nutrient absorption, as well as being a major site of fermentation5,6. The ascending and transverse colons are thought to absorb the remaining water and nutrients, leading to stool compaction before the descending colon is reached. This latter main role is to facilitate the movement of waste material towards the rectum. This tissue compartmentalization translates into different cancer susceptibilities and aetiologies, making colorectal cancer (CRC) a very heterogeneous disease7.
Regular exposure of the colonic epithelium to biomechanical injury, together with an associated obligate high rate of intrinsic cell proliferation, confers an elevated lifetime susceptibility to cancer-causing mutations. Accordingly, CRC is currently the third most diagnosed and second most lethal cancer worldwide, with incidence rates still increasing in many developed countries due to diet and lifestyle changes8. To study and tackle the progression of this disease, the development of physiologically relevant mouse models of advanced colon cancer is of crucial importance. Adult stem cells are known to be major sources of cancer following oncogenic transformation. LGR5 marks resident stem cells throughout the gastrointestinal tract that are responsible for daily renewal of the epithelial lining9. Targeted in vivo dysregulation of the Wnt pathway in gastrointestinal tract stem cells using Lgr5-Cre drivers efficiently drives cancer formation within the mouse small intestine, identifying LGR5+ stem cells as important cancer origins in this tissue9,10. However, the rapid lethality arising from the resulting intestinal tumour load prevents evaluation of LGR5+ colon stem cells as cancer origins and precludes the development and study of more advanced colon tumours in this mouse model. Other CRC models targeting the large intestine have been developed over the years11,12,13,14,15,16. However, to our knowledge, none of the Cre drivers used specifically target the stem cell compartment, precluding the study of cancer originating from and fuelled by this cell population. Identifying markers that selectively enrich for regional colon stem cells and facilitate their selective transformation as an essential prerequisite to evaluating their role in driving cancer and to generate accurate in vivo models of advanced colon cancer would therefore offer clinically relevant mechanistic insights into colon cancer development and deliver valuable pre-clinical screening models.
In this Article, we perform comparative transcriptomics on LGR5+ stem cell populations from different regions of the gastrointestinal tract using LGR5 reporter mouse models to identify and validate markers selectively enriched in LGR5+ colon stem cells. We accordingly identify Nox1 and Npy1r as being highly enriched in LGR5+ colon stem cells in the mouse caecum and middle/distal colorectum, as well as the middle/distal colorectum respectively. Using the mouse models Nox1-2A-Cre recombinase fused with a tamoxifen-inducible estrogen receptor (CreERT2), Nox1-2A-enhanced green fluorescence protein (eGFP) and Npy1r-eGFP-internal ribosome entry site (IRES)-CreERT2, we confirmed by lineage tracing and organoid initiation assays that cells positive for NADPH oxidase 1 (NOX1+) and neuropeptide Y receptor Y1 (NPY1R+) are bona fide stem cells capable of self-renewal and differentiation into both absorptive and secretory cell lineages in the colon. Crucially, we demonstrate that the Nox1-2A-CreERT2 and Npy1r-eGFP-IRES-CreERT2 drivers are capable of selectively initiating cancer in different regions of the colon following Wnt pathway hyperactivation, with minimal to no transformation observed in the small intestine. These observations identify colon stem cells as major cells of cancer origin throughout the colon in mice. Importantly, we also demonstrate that combined activation of Wnt signalling, oncogenic Kras and loss of p53 function in these stem cell compartments results in the formation of more advanced, invasive cancers. These targeted Cre drivers will be invaluable for accurately modelling advanced cancer within different regions of the colon by incorporating prevalent mutations to deliver clinically relevant mechanistic insights into regional cancer progression.
Results
NOX1 selectively enriches for LGR5-expressing stem cells in the colon
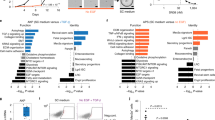
To identify markers specifically enriched in colonic stem cells, we profiled the transcriptome of fluorescence-activated cell sorting (FACS)-isolated GFPhigh and GFPneg cell populations from the small intestine and colon of our non-variegated Lgr5-2A-eGFP reporter mice17 (Fig. 1a). Comparison of their respective GFPhigh and GFPneg transcriptomes revealed the signature of the LGR5-expressing stem cells in each region. As expected, there was a significant overlap with the previous colon LGR5+ stem cell signature generated using Lgr5-eGFP-IRES-CreERT2 reporter mice, including many Wnt target genes18 (Extended Data Fig. 1a). The LGR5+ stem cell signature in the colon overlapped with a published mouse CD44+ colon stem cell signature19, including validated stem cell markers such as achaete-scute family BHLH transcription factor 2 (ASCL2)18 (Supplementary Table 1). We identified the transcriptional signature of GFPhigh stem cells for each tissue by screening it against the corresponding GFPneg cell population. We then screened the list of genes enriched in the colon GFPhigh stem cells against the GFPhigh stem cell transcriptional signature of the small intestine and pylorus20 to identify genes selectively upregulated in colonic stem cells (Fig. 1b and Supplementary Table 2). One of the most highly upregulated targets was Nox1, which is known to be enriched in the distal gastrointestinal tract21,22 (Fig. 1c). Nevertheless, a detailed characterization of the endogenous NOX1 expression pattern and functional evaluation of the stem potential of NOX1-expressing cells within the colon has not yet been performed.
a, Single cells from Lgr5-2A-eGFP mouse colon and small intestine crypts were sorted by FACS according to their level of GFP expression. A published microarray performed in the pylorus using the Lgr5-eGFP-DTR mouse line (in which eGFP is fused to the diphtheria toxin receptor) was used as the pylorus stem cell transcriptomic signature20. From each tissue, GFP+ and GFP− cell populations were collected for transcriptomic analysis. b, The LGR5+ stem cell signature was generated by comparing the gene expression profiles of the GFP+ and GFP− populations for each tissue. These lists of genes were then screened against both the small intestine and pylorus transcriptomic signatures. The UpSet plot represents the common enriched genes in LGR5+ cells between tissues, as well as the enriched genes in LGR5+ cells exclusive to each tissue (the enriched gene number for colonic LGR5+ cells is highlighted in green). c, Nox1 was identified as one of the most enriched genes in the colon stem cell compartment relative to all analysed upper gastrointestinal tract tissues (highlighted in green). d, Colon and small intestine tissues were separated into distinct anatomical parts (proximal, middle and distal small intestine and proximal and distal colon). An LGR5+ stem cell signature was generated for each tissue using the same method as in a. e, UpSet plot representing the common enriched genes in LGR5+ cells between tissues, as well as the enriched genes in LGR5+ cells exclusive to each tissue (the distal colon only is highlighted in red and the proximal colon only is highlighted in green). f, Npy1r was identified as one of the most enriched genes in the distal colon stem cell compartment relative to all analysed upper gastrointestinal tract tissues (highlighted in red). CD, distal colon; CP, proximal colon; CPM, counts per million; FC, fold change; GI, gastrointestinal; SI, small intestine; SID, distal small intestine; SIM, middle small intestine; SIP, proximal small intestine.
NPY1R selectively enriches for LGR5-expressing stem cells in the middle and distal colon
To identify a more regionally restricted colon-specific stem cell marker, we conducted a new series of RNA sequencing (RNA-seq) analyses. Working with Lgr5-2A-eGFP mice, we separated the small intestine into three regions to mimic the anatomical separation of this tissue (that is, the duodenum, jejunum and ileum) and divided the colon into two regions (proximal and distal) (Fig. 1d). We then identified the gene signature for each region by comparing the GFPhigh and GFPneg transcriptomes (Supplementary Tables 3 and 4). Finally, we compared the list of genes enriched in the proximal or distal colon against the GFPhigh stem cell transcriptional signature of the small intestine (all regions) and pylorus. We also compared the list of genes enriched in the distal colon against that of the proximal colon and vice versa to isolate a specific gene signature for each region of the colon (Fig. 1e and Supplementary Tables 5 and 6). Although we failed to validate via quantitative PCR (qPCR) and RNAscope a robustly expressed, selective marker of LGR5+ stem cells in the proximal colon (Extended Data Fig. 1b and Supplementary Table 5), we identified Npy1r as one of the most highly enriched genes in the LGR5+ population of the distal colon (Fig. 1f and Supplementary Table 6). NPY1R is known to be expressed in a subset of enteroendocrine cells in the gastrointestinal tract, but no stem cell enrichment has been described23.
NOX1 is enriched in stem cell populations along the colon, whereas NPY1R is restricted to distal colon stem cells
To precisely define which lineages express NOX1 and NPY1R, we performed single-cell RNA-seq (scRNA-seq) on the epithelial fraction of the small intestine and colon. The small intestine was separated into three parts that were identical to the ones used for RNA-seq analysis (Fig. 1d). The colon was separated into four parts: the caecum and the proximal, middle and distal colon. Epithelial fractions were separated for each tissue the and cells were dissociated and embedded into chromium beads before RNA-seq. Using the shared nearest neighbour (SNN) modularity optimization-based clustering algorithm (implemented in Seurat), we identified 15, 14, 11 and 13 clusters for the caecum and proximal, middle and distal colon epithelium, respectively. Consensus lineage marker expression was plotted for each cluster of each tissue compartment (Fig. 2a) to assign clusters to their respective lineages (Fig. 2b). We then mapped LGR5, NOX1 and NPY1R expression distribution on uniform manifold approximation and projection (UMAP) plots generated for each tissue (Fig. 2c) against the stem cell clusters delineated according to their assigned lineage identities (Fig. 2b). We found that NOX1 was expressed across many clusters at a very low level but highly upregulated within the stem cell cluster for all parts of the colonic epithelium (Fig. 2c), in line with our RNA-seq data. In contrast, NPY1R-expressing cells overlapped with the stem cell cluster specifically in the middle and distal colon regions, with a higher expression in the distal colon, but not in the caecum or proximal colon (Fig. 2c). As was previously shown23, a subpopulation of NPY1R+ cells was found in the chromogranin A (CHGA+) enteroendocrine cell cluster of the colon (Fig. 2c and Extended Data Fig. 1c). Interestingly, we also found a subpopulation of NPY1R-expressing cells in the tuft cell cluster in the distal colon (Fig. 2c).
a, Heatmap representing the level of expression of identified key lineage markers for each cluster in the caecum and proximal, middle and distal colon. b, UMAP plots representing all of the clusters identified (left) before being reassigned to different lineage identities using the expression heatmap in a (right). c, UMAP plots representing the distribution of LGR5+ (top), NOX1+ (middle) and NPY1R+ cells (bottom) for each colonic region (caecum and proximal, middle and distal colon). The stem cell (SC) cluster is delineated by a blue dotted line, the enteroendocrine (EE) cluster is delineated by a brown dashed line and the tuft cluster is delineated by a purple dashed line, according to the expression of consensus lineage markers as defined in b. Assays were performed on one biological sample per tissue region. CM, middle colon; DCS, deep crypt secretory.
We quantified the total number of LGR5+ cells for each cluster and their expression status with respect to NOX1 and NPY1R expression (Extended Data Fig. 1d). We found that in the caecum 75.75% of the LGR5+ stem cells co-expressed NOX1. Surprisingly, NOX1 was found to be co-expressed in only 29.15% of LGR5+ stem cells in the proximal colon. This percentage increased towards the middle and distal part of the colon, with 84.52 and 77.73% of LGR5+ stem cells co-expressing NOX1, respectively (Extended Data Fig. 1d). When looking at NPY1R expression in LGR5+ stem cells, we showed that the percentage of co-expression increased from the middle to the distal colon (27.61 and 62.28%, respectively) and that the majority of the LGR5+ stem cells (52.73%) co-expressed both NOX1 and NPY1R (Extended Data Fig. 1d). When looking at NOX1+ and NPY1R+ cells for each cluster, we also found that the majority of these cells were within the stem cell cluster (Extended Data Fig. 1e,f). For the caecum, we found that 66.08% of NOX1+ stem cells expressed LGR5. This percentage remained constant in the proximal colon (65.22%) and distal colon (67.06%), but decreased in the middle colon (42.62%) (Extended Data Fig. 1e). NPY1R+ stem cells were found to be present predominantly in the distal colon, with most co-expressing LGR5 and NOX1 (Extended Data Fig. 1f). Overall, these data show that NOX1 is expressed in the stem cell compartment of the entire large intestine, with the highest overlap in the caecum and distal colon (Extended Data Fig. 1d,e), whereas NPY1R expression is highly restricted to the distal colon stem cell compartment (Extended Data Fig. 1d,f).
For our small intestine scRNA-seq dataset, we again used the SNN modularity optimization-based clustering algorithm to identify 14, 15 and 16 clusters for the proximal, middle and distal small intestine epithelium, respectively. We plotted the key lineage marker expression for each cluster of each region (Extended Data Fig. 2a) in order to assign each cluster to its respective lineage identity on a UMAP plot (Extended Data Fig. 2b). We found NOX1 expression to be distributed at extremely low, almost undetectable levels throughout all clusters of each part of the small intestine (Extended Data Fig. 2c). In contrast, we did not detect any NPY1R+ cells in all regions of the small intestine except a subpopulation within the enteroendocrine cluster, which presented a very low level of NPY1R expression (Extended Data Fig. 2c). Taken together, these results demonstrate that NOX1 is expressed in stem cell compartments throughout the colon, with the highest enrichment present within the caecum and middle and distal regions. In contrast, NPY1R is selectively enriched in LGR5+ stem cell compartments of the middle and distal colon, with the highest enrichment in distal regions.
Nox1 expression is restricted to crypt bases of the colon
Next, we validated our in silico transcriptomic analyses by first establishing endogenous Nox1 RNA expression profiles within different regions of the colon using RNAscope. In the caecum and distal colorectum, the Nox1 expression pattern was highly similar to that of Lgr5, with transcripts restricted to crypt bases (Fig. 3a). In the proximal colon, Nox1 expression was similarly restricted to crypt bases but was only detected in a minority of crypts and at notably lower levels than Lgr5 (Fig. 3a). In the middle colon, Nox1 expression extended beyond the Lgr5+ stem cell compartment at the crypt base to encompass presumptive early transit-amplifying cells (Fig. 3a).
a, RNAscope analysis of Lgr5, Nox1 and Npy1r in the different regions of the mouse colon, revealing enrichment of NOX1 and NPY1R at the crypt base of all compartments and the middle and distal colorectum, respectively (n = 5). b,c, Nox1-2A-eGFP (n = 5) (b) and Npy1r-eGFP-IRES-CreERT2 (n = 5) (c) mouse models were generated by incorporating the 2A-eGFP cassette before the stop codon at the Nox1 gene locus and the eGFP-IRES-CreERT2 cassette after the start codon at the Npy1r gene locus. GFP IHC recapitulated endogenous NOX1 and NPY1R expression, as seen in a. GFP expression was also detected in the base of rectal crypts in Npy1r knock-in mice. Scale bars, 25 μm.
Nox1 expression was largely absent from the stomach, duodenum and jejunum (Extended Data Fig. 3a), except for intestinal crypts directly adjacent to Peyer’s patches, where expression was restricted to crypt bases (Extended Data Fig. 3b). Limited Nox1 expression was also detected at crypt bases in the most distal part of the ileum, with robust expression present around the +4 to +6 positions immediately adjacent to the Paneth cell compartment and low levels in a subset of crypt base columnar cells (Extended Data Fig. 3b).
Collectively, these expression analyses corroborate the in silico data, identifying Nox1 as being highly and selectively expressed in Lgr5+ epithelial stem cell compartments throughout the adult colon, with the greatest enrichment present within the caecum and distal colorectum.
Npy1r expression is restricted to crypt bases of the middle and distal colon
In contrast with Nox1, Npy1r expression was absent from the proximal colon and caecum of adult mice (Fig. 3a). In the middle colon, relatively weak Npy1r expression extended beyond the Lgr5+ zone at the crypt base to encompass the lower third of the crypts in a pattern reminiscent of Nox1 expression in this region (Fig. 3a). More robust levels of Npy1r were present exclusively within the Lgr5+ zone at crypt bases throughout the distal colorectum (Fig. 3a). Npy1r expression was absent from epithelia of the small intestine, stomach and bladder, but exhibited robust levels in bladder stroma where Npy1r is known to mark a subset of fibroblast cells24 (Extended Data Fig. 3c,d). Again, these expression analyses aligned well with our in silico data, identifying Npy1r as being highly enriched in Lgr5+ epithelial stem cell compartments predominantly within the distal adult colorectum.
Nox1 and Npy1r knock-in mice faithfully report endogenous expression in the colon
To further characterize the various NOX1- and NPY1R-expressing colon populations, we generated independent eGFP reporter mouse models via targeted integration of either 2A-eGFP or eGFP-IRES-CreERT2 cassettes at the Nox1 or Npy1r locus, respectively (Fig. 3b,c). GFP expression in adult Nox1-2A-eGFP reporter mice faithfully recapitulated endogenous NOX1 expression across all regions of the colon and ileum (Fig. 3b and Extended Data Fig. 4a). Similarly, GFP expression in Npy1r-eGFP-IRES-CreERT2 reporter mice was predominantly restricted to crypt bases of the middle and distal colorectum, recapitulating the endogenous expression pattern (Fig. 3c). Sporadic reporter expression was also observed scattered in stromal tissues throughout the colon and rectum (Fig. 3c) and bladder (Extended Data Fig. 4b).
Next, we sorted the GFPhigh and GFPneg cell populations from the different adult reporter mice by FACS to document the lineage identities of the NOX1+ and NPY1R+ colon populations using established markers via qPCR. For both NOX1-GFPhigh and NPY1R-GFPhigh cell populations, Nox1 and Npy1r, respectively, together with Lgr5 and the stem cell markers SPARC-related modular calcium-binding protein 2 (Smoc2)25 and Ascl2, were robustly enriched compared with in GFPneg populations (Extended Data Fig. 5a), further underscoring their relatively undifferentiated, stem-like identity. Npy1r was not enriched in the caecum, in line with our scRNA-seq and staining data (Extended Data Fig. 5a). Conversely, expression levels of the lineage markers downregulated in adenomas (Dra, also called Slc26a3—a colonocyte marker), Mucin2 (Muc2—a goblet marker) and regenerating family member 4 (Reg4—a deep crypt secretory cell marker) were markedly lower in GFPhigh compared with GFPneg cell populations. The expression of Chga (an enteroendocrine marker) was also relatively low in NOX1+ populations. In contrast, NPY1R+ populations in the distal colon displayed robust ChgA expression (Extended Data Fig. 5a), consistent with observations from scRNA-seq analyses documenting the presence of enteroendocrine cells within this population (Fig. 2 and Extended Data Fig. 1c).
Co-immunofluorescence (co-IF) of GFP with CHGA, MUC2 and DRA across the different regions of the colon from adult NOX1 reporter mice confirmed a lack of co-localization with these mature lineage markers, further highlighting the relatively undifferentiated status of NOX1+ populations (Extended Data Fig. 5b). NPY1R-GFP+ colon populations similarly lacked co-expression of MUC2 or DRA markers, but did partially co-localize with CHGA, confirming that a subset of NPY1R+ cells are enteroendocrine cells (Extended Data Fig. 5c). A subset of NOX1-GFP+ and NPY1R-GFP+ cells overlapped with the proliferation marker Ki-67, highlighting their proliferative status (Extended Data Fig. 5b,c).
NOX1+ and NPY1R+ cells establish long-term self-renewing colonic organoids in vitro
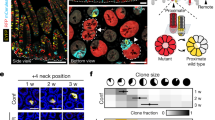
To evaluate the stem cell potential of NOX1-expressing colon cells, we FACS-sorted NOX1-GFPhigh and NOX1-GFPneg cells from the caecum and distal colon of adult Nox1-2A-eGFP reporter mice and assessed their capacity to form long-term organoid cultures in vitro (Fig. 4a). The organoid outgrowth efficacy of plated GFPhigh cells relative to GFPneg cells was 11-fold higher in the caecum and 5.5-fold higher for the distal colon (Fig. 4b). Importantly, the limited numbers of caecum and colon organoids generated from GFPneg cells could not be maintained beyond the first passage, in contrast with GFPhigh cell-derived organoids, which could be cultured long term (more than ten passages) (Fig. 4c).
a, Nox1-2A-eGFP single cells from caecum and distal colon crypts were isolated by FACS according to their GFP expression level. The lines on the plot indicate the boundaries of the gates set for demarcating a GFP-neg and GFP-high population. The x-axis indicates that the GFP signal was measured by excitation with a 488 nm laser and emission collected by a 530/40 nm filter. b, Caecum (n = 3; 3.2633 ± 0.1335%) and colon (n = 3; 3.9233 ± 0.3398%) organoid outgrowth efficiencies were calculated as the number of organoids formed at passage 0 (P0) relative to the total number of cells seeded. Only GFP+ cells had the potential to grow organoids. c, Representative images of newly established caecum (top) and distal colon organoids (bottom) at P0 and P10. GFP+ cell-derived organoids were passaged for more than ten passages. d, Npy1r-eGFP-IRES-CreERT2 single cells from the distal colon were isolated by FACS according to their GFP expression level. The lines on the plot indicate the boundaries of the gates set for demarcating a GFP-neg, GFP-mid and GFP-high population. The x-axis indicates that the GFP signal was measured by excitation with a 488 nm laser and emission collected by a 530/40 nm filter. e, The distal colon organoid outgrowth efficiencies of GFP− and GFP+ cells showed that only GFP+ cells had the potential to grow organoids (n = 6; 5.6166 ± 0.8734%). f, Representative images of distal colon organoids at P0 and P10. GFP+ cell-derived organoids were passaged for more than ten passages. g, Schematic of the organoid differentiation protocol used to culture Matrigel-seeded cells, first for 2 d in organoid growth medium (OGM) before switching to organoid differentiation medium (ODM) once the organoids had been established for 3 (distal colon) or 5 d (caecum). h,i, qPCR analysis of Nox1-2A-eGFP-derived caecum (h) and distal colon (i) organoids, showing a decrease in the expression of Nox1 (caecum = 0.1253 ± 7.623 × 10−3; colon = 0.02760 ± 6.952 × 10−4) and Lgr5 (caecum = 0.1336 ± 5.044 × 10−3; colon = 0.03647 ± 9.903 × 10−4) and an increase in the expression of the lineage markers Dra (caecum = 97.575 ± 15.55; colon = 7.020 ± 0,5976), Muc2 (caecum = 3.3936 ± 0.1463; colon = 5.765 ± 0.5768) and ChgA (caecum = 4.073 ± 0.2047; colon = 5.147 ± 0.1707) after differentiation (n = 3). j, qPCR analysis of Npy1r-eGFP-IRES-CreERT2-derived distal colon organoids showing a decrease in the expression of Lgr5 (0.112 ± 9.644 × 10−3) and an increase in the expression of the lineage markers Dra (23.9223 ± 1.908), Muc2 (2.8366 ± 0.1787) and ChgA (3.4676 ± 0.1252) after differentiation (n = 3). Npy1r expression was unchanged after differentiation (0.8083 ± 0.1501). Statistical significance in b, e and h–j was determined by two-tailed t-test and the data are presented as means ± s.e.m. Scale bars, 150 μm. FSC, forward scatter.
We also FACS sorted NPY1R-GFPhigh and NPY1R-GFPneg cells from the distal colon of adult Npy1r-eGFP-IRES-CreERT2 reporter mice (Fig. 4d). The organoid outgrowth efficacy of plated GFPhigh cells relative to GFPneg cells was 5.5-fold higher (Fig. 4e). Again, the limited number of GFPneg cell-derived organoids could not be maintained beyond the first passage, in contrast with the GFPhigh cell-derived organoids that could be passaged long term (Fig. 4f).
To formally document the in vitro multipotency of the plated NOX1+ and NPY1R+ cells, we performed a differentiation assay. Organoids derived from either NOX1+ or NPY1R+ cells were cultured in growth medium for 2 d before switching to a differentiation medium for 3 and 5 d for the distal colon and caecum, respectively (Fig. 4g). qPCR analyses on NOX1+ cell-derived caecum and distal colon organoids documented a major decrease in both Nox1 and Lgr5 expression and a concomitant increase in differentiated lineage marker expression (Fig. 4h,i). NPY1R+ cell-derived distal colon organoids similarly displayed a marked increase in differentiated lineage marker expression, with only a modest accompanying decrease in Npy1r, probably due to the presence of Npy1r-expressing enteroendocrine cells (Fig. 4j). Collectively these results identify NOX1+ populations from both the caecum and distal colon and the NPY1R+ population from the distal colon as being highly enriched for cells with robust stem cell potential in vitro.
NOX1 marks self-renewing, multipotent colon stem cell populations in vivo
To functionally evaluate the endogenous stem cell identity of NOX1-expressing cells within the adult colonic epithelium, we generated a Nox1-2A-CreERT2/Rosa26-tdTomato (tdTom) mouse model to track the contribution of endogenous NOX1-expressing cells to homeostatic epithelial renewal via in vivo lineage tracing (Fig. 5a). At 24 h after tamoxifen induction, cells at the base of the crypts of the colon expressed tdTom, with only a subset of labelled crypts in the proximal colon, recapitulating the expression profiles observed via RNAscope analyses and the eGFP reporter model (Fig. 5b). One week after induction, tdTom-labelled progeny had expanded to encompass most of the crypts (Fig. 5b). One month after tamoxifen induction, the vast majority of crypts and the associated surface epithelium in the caecum (97.7%), middle (91.7%) and distal colon (92.5%) and rectum (75%), together with scattered crypts in the proximal colon, were entirely tdTom positive (Fig. 5b and Extended Data Fig. 6a). Co-IF analyses of tdTom with major cell lineage markers confirmed overlapping expression in tracing units across all regions of the colon (Fig. 5c). Collectively, these observations identify NOX1+ populations as harbouring self-renewing, multipotent stem cells contributing to epithelial homeostasis throughout the colon.
a, A Nox1-2A-CreERT2 mouse model was generated by incorporating the 2A-CreERT2 cassette before the stop codon at the Nox1 gene locus. Mice were crossed with the Rosa26-LSL-tdTom mouse line. b, Mice were injected with 3 mg tamoxifen per 30 g body weight (3 mg/30 g) and analysed at 24 h (n = 3), 1 week (n = 3) and 1 month (n = 3) after induction. RFP IHC showed lineage tracing of NOX1-expressing cells and their cell progeny. c, Mice were injected at a lower tamoxifen dose (0.1 mg per 30 g body weight) and harvested at 1 month (n = 3). Co-IF of RFP and the lineage markers MUC2, CHGA and DRA showed that all RFP+ traced units expressed major lineage markers. Scale bars, 25 μm.
We further explored the clonal dynamics of lineage tracing using the Nox1-2A-CreERT2 driver by administering a low dose of tamoxifen (0.1 mg per 30 g bodyweight) to initiate recombination at low frequency. This tamoxifen dose is sufficient to activate tdTom expression within single cells at colonic crypt bases after 24 h in both the middle and distal colon (top panel of Extended Data Fig. 6b). As expected, tracing units initiated from these single NOX1+ cells had expanded upwards from the crypt base after 1 week (middle panel of Extended Data Fig. 6b). A subset of the tracing units further expanded to encompass entire crypts by 1 month (bottom panel of Extended Data Fig. 6b), confirming that individual NOX1-expressing stem cells have the potential to populate entire colonic crypts during epithelial renewal. With this low level of induction, the proportion of crypts containing tdTom+ cells rapidly decreased over time, from 95.66 ± 3.44 and 87.33 ± 6.15% of crypts at 24 h to 48.44 ± 14.37 and 44 ± 8.88% of crypts after 1 week and 31.11 ± 9.11 and 34.44 ± 7.40% of crypts after 1 month in the middle and distal colon, respectively (Extended Data Fig. 6c). The decreases in the number of tdTom+ crypts 24 h and 1 month after induction in the middle and distal colon were statistically significant (P < 0.0001), probably reflecting known stem cell competition dynamics between labelled and non-labelled colonic stem cells in these crypts26,27,28. In contrast, using the same low tamoxifen dose, we found that a larger pool of NOX1-expressing cells were labelled at crypt bases of the caecum after 24 h (Extended Data Fig. 6b), resulting in a higher frequency of retained lineage tracing after 1 month (70.4 ± 10.98% tdTom+ caecum crypts after 24 h and 61.33 ± 9% after 1 month; P = 0.0674) (Extended Data Fig. 6c). Thus, NOX1-expressing cells present distinct lineage tracing dynamics in different regions of the colon, with more efficient recombination and retention of clones within the caecum.
In the small intestine, long-term tracing was only observed in the distal ileum and crypts adjacent to the Peyer’s patches (Extended Data Fig. 7a,b), in line with the endogenous NOX1 expression observed within the LGR5+ stem cell compartments of those regions (Extended Data Figs. 3b and 4a). Following induction with 3 mg tamoxifen per 30 g bodyweight, lineage tracing in the ileum originated predominantly from presumptive early transit-amplifying cells immediately adjacent to the Paneth cell compartment, although tdTom expression was also observed in limited numbers of crypt base columnar cells interspersed with Paneth cells at the crypt base after 24 h (Extended Data Fig. 7a). Accordingly, limited numbers of tdTom+ ribbons spanning crypt–villus units present in the distal ileum after 1 week were retained after 1 month (Extended Data Fig. 7a). In contrast, multiple tdTom+ cells were observed at the base of crypts adjacent to Peyer’s patches after 24 h (Extended Data Fig. 7b). After 1 week, tracing units had expanded along the entire crypt–villus axis and the follicle-associated surface epithelium (Extended Data Fig. 7b). By 1 month, tracing units had further expanded to completely encompass the Peyer’s patch-associated crypt–villus epithelium and the follicle lining epithelium (Extended Data Fig. 7b). These data highlight Nox1-2A-CreERT2 as being a highly selective colonic stem cell driver that readily facilitates recombination throughout the colon, along with accompanying sporadic recombination within the ileum.
NPY1R marks self-renewing, multipotent distal colon stem cell populations in vivo
To evaluate the endogenous stem cell identity of NPY1R+ colon cells, we performed in vivo lineage tracing using Npy1r-eGFP-IRES-CreERT2, tdTom mice under homeostatic conditions (Fig. 6a). Due to the relatively low endogenous expression levels of NPY1R in the colon, we increased the tamoxifen dose to 4 mg per 30 g bodyweight to ensure robust recombination levels. At 24 h post-induction, single tdTom+ cells were visible at the crypt bases within the middle colon, whereas a higher recombination frequency was achieved throughout the distal colon and rectum, with multiple tdTom+ cells present at most crypt bases (Fig. 6b). After 1 week, basal tdTom+ tracing units had expanded to reach the surface epithelium in multiple crypts in both the middle and distal colon and rectum, albeit at a higher frequency within more distal and rectal regions (Fig. 6b). The tracing frequency in the distal colon and rectum was maintained after 1 month, with 74.8% of colon crypts and associated surface epithelium displaying uniform tdTom expression and 81.8% of rectum crypts retaining tdTom labelling. Completely traced crypts were also present in the middle colon at this time point, albeit at a reduced frequency (22.7%) reflecting the lower induction efficacy in this region (Fig. 6b and Extended Data Fig. 8a). As expected, recombination within the crypts of the proximal colon was exclusively initiated within infrequent presumptive enteroendocrine cells at 24 h, but this was not retained at later time points (Fig. 6b).
a, Npy1r-eGFP-IRES-CreERT2 mice were bred with a Rosa26-LSL-tdTom mouse line. b, Mice were injected with 4 mg tamoxifen per 30 g bodyweight and harvested at 24 h (n = 3), 1 week (n = 3) or 1 month (n = 3) after injection. At 24 h, an RFP signal was detected in the epithelium of the proximal colon (see magnified insert). RFP was detected at the base of the crypt of the middle and distal colon, as well as the rectum. 1 week after induction, the RFP signal was lost in the proximal colon, whereas it spread from the base of the crypt in the middle and distal colon and the rectum. At 1 month, only the middle colon (at a lower frequency), distal colon and rectal crypts retained the RFP signal. c, Mice were injected with 3 mg tamoxifen per 30 g bodyweight and harvested after 1 month (n = 3). Co-IF of RFP and the lineage markers MUC2, CHGA and DRA showed that all RFP+ traced units expressed major lineage markers. Scale bars, 25 μm.
To formally document the multipotency of NPY1R+ colon cells, we performed red fluorescent protein (RFP) co-IF with CHGA, DRA and MUC2. Each lineage marker could be detected within tdTom+ tracing units present in the middle and distal colon, confirming that NPY1R+ cells can generate the various functional lineages of the colonic epithelium (Fig. 6c). We therefore conclude that NPY1R+ crypt-base-resident populations in the middle and distal colorectum are highly enriched for long-term self-renewing, multipotent cells contributing to homeostatic epithelial renewal.
We additionally performed lineage tracing in the bladder to assess the behaviour of resident NPY1R+ populations. After 24 h induction, tdTom+ cells were detected within the stroma of the epithelium. This labelling was retained over time after 1 week and 1 month of tracing, indicating that NPY1R marks long-lived stromal cells in the bladder (Extended Data Fig. 8b). We performed co-IF staining of RFP with lineage markers. RFP+ cells failed to co-localize with the epithelium marker E-cadherin and the smooth muscle marker alpha smooth muscle actin. In contrast, RFP+ cells were uniformly positive for the mesenchyme marker vimentin, confirming the stromal identity of NPY1R+ bladder cells (Extended Data Fig. 8c).
Targeted mutation of the genome of NOX1+ stem cells drives cancer initiation predominantly in the caecum
LGR5+ stem cells in the small intestine are major cells of cancer origin following targeted dysregulation of Wnt signalling in vivo using Lgr5-CreERT2 drivers10. The ensuing rapid tumour growth in the small intestine causes rapid lethality, precluding rigorous evaluation of LGR5+ stem cell populations as cancer origins in the different regions of the colon. Having established the selective enrichment of NOX1 in stem cells within the caecum and middle and distal colon, we reasoned that the Nox1-2A-CreERT2 driver should facilitate selective targeting of oncogenic mutations to the genome of colon stem cells as an essential prerequisite to evaluating their role in driving colon cancer initiation. Hyperactivation of Wnt signalling via loss-of-function mutation of APC is a highly prevalent early event in human CRC29. We therefore generated a Nox1-2A-CreERT2;Apcfl/fl mouse model that incorporates a tdTom reporter allele to facilitate the visualization of any tumour formation in the gastrointestinal tract following targeted transformation of NOX1+ stem cells (Fig. 7a). Non-induced adult mice were healthy, displayed very low levels of spontaneous recombination (as evidenced by <1 in 500 tdTom+ colon crypts) and presented no evidence of Wnt pathway hyperactivation with the absence of β-catenin accumulation within all regions of the colonic epithelium (Extended Data Fig. 9a). One month after tamoxifen administration, nucleocytoplasmic accumulation of β-catenin (β-cathi) was apparent throughout the caecum and within discrete regions of the middle and distal colon (Fig. 7a). Co-IF for Ki-67 and β-catenin revealed that a subset of the β-cathi cells were actively proliferating in the different regions (Fig. 7b). Compared with control tissues, these Wnt-hyperactivated crypts were highly disorganized and displayed prominent signs of hyperplasia (Fig. 7a,b). In line with the higher efficiency of recombination within the caecum (Extended Data Fig. 6b,c), the caecum presented more polyps and a markedly higher proportion of crypts displaying nucleocytoplasmic β-catenin accumulation compared with crypts present in the middle and distal colon (Fig. 7a). To a lesser extent, we note an accumulation of β-catenin in the Peyer’s patches of the small intestine within small proliferative cell clusters (Extended Data Fig. 9b,c). Crucially, unlike previous LGR5-driven cancer models in which small intestinal tumours rapidly develop10, we did not detect any nucleocytoplasmic accumulation of β-catenin within the ileum, even after 1 month of induction (Extended Data Fig. 9b), highlighting the selectivity of the Nox1-2A-CreERT2 driver for initiating cancers within the colon as an essential prerequisite to modelling more advanced cancers in the caecum and distal colon via the incorporation of additional, physiologically relevant mutations.
a, The Nox1-2A-CreERT2 driver mouse line was crossed with the Rosa26-LSL-tdTom and Apcfl/fl mouse lines. Mice were bred to homozygosity. Mice were injected twice 2 d apart with 4 mg tamoxifen per 30 g bodyweight and harvested 1 month after injection (n = 4). β-catenin IHC showed nucleocytoplasmic accumulation in the caecum and middle and distal colon 1 month after induction. b, Co-IF for β-catenin and the proliferation marker Ki-67 showed hyperproliferation in the crypts of the caecum and middle and distal colon following Apc deletion (n = 4). c, Nox1-2A-CreERT2, Apcfl/fl, Rosa26-LSL-tdTom mice were bred with the inducible LSL-KrasG12D strain. LSL-KrasG12D was kept heterozygotic. Mice were injected with 1 mg tamoxifen per 30 g bodyweight and harvested 1 month later (n = 3). β-catenin IHC showed nucleocytoplasmic accumulation throughout the caecum epithelium, with hyperactivation of Kras shown by an increase of pMapk. Only one polyp over four different colons was detected with nucleocytoplasmic β-catenin accumulation and pMapk hyperactivation. The rest of the colon presented with normal membranous β-catenin with no nucleocytoplasmic accumulation. d, A p53fl/fl allele was incorporated into the Nox1-2A-CreERT2, Apcfl/+, LSL-KrasG12D mouse model. Mice were injected with 2 mg tamoxifen per 30 g bodyweight and harvested 3 months after induction (n = 4). Tumours in the caecum presented with adenocarcinoma features with high levels of β-catenin accumulation, validating the loss of Apc heterozygosity. E-cadherin+ cells had invaded the submucosa layer until the muscularis for the most advanced stage. Invading tumours were hyperactivated for KRAS (pMAPK+) and devoid of p53 expression, highlighting efficient allelic recombination. Scale bars, 25 μm. D, day.
Given the highly efficient recombination achieved in the caecum relative to the distal colon, we reasoned that reducing the tamoxifen induction dose would enable selective targeting of mutations affecting the caecum epithelium to facilitate the generation of a caecum-specific cancer model. As caecum cancer presents the highest rate of Kras mutation compared with other forms of CRC30,31, we incorporated a conditional KrasG12D allele into our Nox1-2A-CreERT2, Apcfl/fl mouse model (Fig. 7c). Following low dose tamoxifen induction of adult mice at 1 mg per 30 g bodyweight, we noted a major decrease of β-cathi polyps in the distal colon (one polyp was detected over four independent mouse colons), with no accompanying hyperactivation of phosphorylated MAPK (pMAPK). In contrast, the entire caecum epithelium harboured β-cathi polyps displaying robust hyperactivation of pMAPK, indicating a high recombination efficacy for both Apc and Kras alleles in the tissue (Fig. 7c). No β-cathi polyps were visible in the small intestine for both the ileum and Peyer’s patches, and pMAPK activity was confined to the crypt base columnar cells and transit-amplifying compartment, as seen in non-induced mice (Extended Data Fig. 9d).
To further demonstrate the value of using the Nox1-2A-CreERT2 driver to generate more advanced cancers, we incorporated the conditional knockout mutant allele p53fl/fl into our model. We reasoned that incorporating a single floxed Apc allele to this compound mutant model would ensure a limited tumour load resulting from sporadic loss of heterozygosity of the second wild-type Apc allele, thereby ensuring a lifespan compatible with cancer progression, as previously shown32. Indeed, induction of the resulting Nox1-ATK mice resulted in the formation of an average of four to six large tumours in the caecum after 3 months, including advanced tumours independently characterized by a certified animal pathologist as being T2-stage adenocarcinomas undergoing robust invasion of the underlying submucosal layers. Immunohistochemistry (IHC) analyses confirmed successful recombination of the three conditional alleles in these tumours and loss of heterozygosity of Apc, as highlighted by β-catenin accumulation, pMAPK expression and loss of p53 expression (Fig. 7d). These results collectively identify NOX1+ stem cells in the caecum and distal colon as key sources of cancer following in vivo mutation. Importantly, lower tamoxifen induction doses facilitate highly efficient targeting of conditional mutations affecting the caecum to selectively drive advanced cancer in this region.
Targeted mutation of the genome of NPY1R+ stem cells drives cancer initiation in the distal colon
The well-established gene signature for distal CRC involves the sequential mutation of Apc, Kras, Trp53 and Smad4 alleles33,34. First, we incorporated the conditional Apcfl/fl and conditional KrasG12D alleles by crossing with our Npy1r-eGFP-IRES-CreERT2, LoxP-Stop-LoxP (LSL)-tdTom mice (Fig. 8a). Non-induced mice showed no ectopic Cre activation in the colon (Extended Data Fig. 10a). Because of the low level of NPY1R expression, we increased the tamoxifen dosing regimen to three consecutive doses of 4 mg per 30 g bodyweight every 2 d. Due to the relatively slow growth of distal colon tumours compared with cancers originating in the caecum (Fig. 7b), mice were harvested at an extended timepoint of 8 weeks after induction to allow sufficient growth for detection. Nucleocytoplasmic accumulation of β-catenin was readily apparent only in the rectum, probably reflecting the relatively high levels of NPY1R-driven Cre expression levels in this region (Fig. 8b). pMAPK hyperactivation was detected throughout the entire rectum epithelium (Fig. 8b). Co-IF imaging of β-catenin and pMAPK showed that β-catenin-accumulating cells were hyperactivated for pMAPK (Fig. 8b). Co-IF imaging with the Ki-67 marker revealed that these Wnt-hyperactivated cells were highly proliferative (Fig. 8b). No β-catenin accumulation was observed in the bladder after 8 weeks of induction (Extended Data Fig. 10b).
a, The Npy1r-eGFP-IRES-CreERT2 driver mouse line was crossed with the Rosa26-LSL-tdTom, Apcfl/fl and LSL-KrasG12D mouse lines. Mice were bred to homozygosity and LSL-KrasG12D was kept heterozygotic. Mice were injected three times 2 d apart with 4 mg tamoxifen per 30 g bodyweight and harvested 8 weeks after injection (n = 3). b, β-catenin and pMAPK IHC showed localized nucleocytoplasmic accumulation in the rectum and pMAPK hyperactivation throughout the rectal epithelium. Co-IF for β-catenin and pMAPK showed co-localization of nucleocytoplasmic β-catenin and pMAPK hyperactivation. Co-IF for β-catenin and Ki-67 showed hyperproliferation in the crypts accumulating β-catenin following Apc deletion and Kras hyperactivation (n = 3). c, The p53fl/fl allele was incorporated into our mouse model. Mice were injected with three consecutive tamoxifen doses at 4 mg per 30 g body weight at 2-d intervals and harvested 3 months after induction (n = 1). Tumours presented with adenocarcinoma features, with β-catenin-accumulating E-cadherin+ epithelial cells invading the subepithelial layer. These cells were hyperactivated for pMAPK and devoid of p53. Scale bars, 25 μm.
Finally, we incorporated the p53fl/fl conditional knockout allele into the Npy1r-eGFP-IRES-CreERT2hom, Apcfl/fl, KrasG12D/+ mouse model. Npy1r-eGFP-IRES-CreERT2 was used as a homozygote to ensure sufficient Cre expression for the necessary multi-allelic recombination. Although the Npy1r-CreERT2 homozygote is effectively a knockout, Npy1r-knockout mice were healthy and fertile, as previously shown35. The colon was healthy and did not present any visible defects. At 3 months following tamoxifen induction, mice presented with a high tumour load in the rectal area with frequent rectal prolapses. Tumour features were independently characterized by a certified animal pathologist as adenocarcinomas displaying an onset of invasion through the submucosa layers (Fig. 8c). Thus, NPY1R+ stem cells in the rectum serve as important sources of cancer formation following dysregulation of Wnt signalling in vivo. Incorporation of relevant compound conditional mutant alleles to the mouse validates the Npy1r-Cre driver as a powerful tool with which to model more advanced stages of rectal cancer originating from stem cells.
Discussion
In this study, we identify NOX1 and NPY1R as markers enriched in regionally distinct epithelial stem cell compartments within the colon. We demonstrate that NOX1+ and NPY1R+ cells are resident colonic stem cells that display properties of self-renewal and multipotency both in vivo and in vitro, expanding the limited toolkit of stem cell markers currently available for the study of this cell population. Indeed, colonic stem cells have to date been identified using the markers LGR5, SMOC2, ASCL2 and CD44, which also label the crypt base columnar stem cells of the small intestine, among other tissues18,19,25. Moreover, existing stem cell markers are expressed across all regions of the colon, greatly limiting their utility for identifying and characterizing the roles of diverse stem cell populations residing in tissue locations along the proximal-to-distal axis, which are known to have clear underlying differences in morphology, epithelial turnover and functions2,3,4,7. Our work therefore addresses this gap by identifying proximal (caecum)-enriched and distal-enriched colonic stem cell populations labelled, respectively, by NOX1 and NPY1R, allowing an in-depth evaluation of these region-specific populations in driving homeostasis, regeneration and disease.
We have previously employed Lgr5-CreERT2 mouse models to identify LGR5+ stem cell populations in the small intestine as key cells of origin of Wnt-driven intestinal cancer10. However, the rapid lethality arising from the resulting tumour load in the small intestine precluded an evaluation of colon LGR5+ stem cells as potential cancer origins using this model. Using colon-specific Nox1-2A-CreERT2 and Npy1r-eGFP-IRES-CreERT2 models that facilitate selective targeting of physiologically relevant conditional mutations to the genome of colon stem cells in the caecum or distal colorectum, respectively, we now formally document regional colon stem cell pools as important cancer origins. Crucially, by modulating the tamoxifen dosage administered to our NOX1 and NPY1R models and incorporating relevant sets of conditional mutant alleles known to drive human CRC, we can selectively initiate regional CRC formation that advances to invasive adenocarcinoma stages. These results highlight the potential of these stem cell Cre drivers for accurately modelling advanced CRC across different regions of the colon.
The value of such regional CRC models has emerged in part due to recent profiling efforts highlighting that colon cancer is a much more heterogeneous disease than was previously appreciated, with cancer aetiology, progression and treatment response dictated by their site of origin within the colon36,37,38. Indeed, the underlying mutation signatures differ markedly between proximal and distal colon cancers30,31, and various factors, including the composition of the resident microbiome and local biomechanical inputs, are now recognized to influence cancer development and treatment responses39,40. At the same time, both caecum and rectal cancers present differing, yet highly debilitating, disease phenotypes and outcomes: caecum tumours are typically detected at advanced stages, accompanied by a high propensity for lymph node metastasis that ultimately results in poor prognosis41,42, whereas rectum cancers, which account for 40% of all CRC cases, can be detected earlier but continue to reflect a high incidence and mortality8, particularly in younger populations43. There is therefore an urgent need to develop more accurate CRC models that recapitulate intrinsic regional heterogeneities to deliver clinically relevant mechanistic insights that can underpin the development of more accurate diagnostic biomarkers and therapeutic targets in tackling this disease. We anticipate that our identification of NOX1 and NPY1R as region-specific colorectal stem cell markers, along with the generation of mouse models that can drive cancer initiation selectively in distinct regions of the colon, will serve as important pre-clinical models guiding these therapeutic efforts.
Methods
Mice
Mouse lines
For Nox1-2A-eGFP and Nox1-2A-CreERT2, the cassettes were inserted immediately before the stop codon of the Nox1 gene locus by homologous recombination in embryonic stem cells, thereby preserving the intact protein-coding region and endogenous expression of the genes. For Npy1r-eGFP-IRES-CreERT2, the cassettes were inserted immediately after the start codon of the Npy1r gene locus by homologous recombination in embryonic stem cells, thus eGFP and CreERT2 expression was under the direct control of the Npy1r locus promoter. Rosa26-LSL-tdTom (Ai14) (007914; The Jackson Laboratory) and LSL-KrasG12D (019104; The Jackson Laboratory) mice were obtained from The Jackson Laboratory. Lgr5-2A-eGFP17, Apcfl/fl (1857966; Mouse Genome Informatics)44 and Trp53fl/fl (ref. 45) mice have been described previously. All of the lines were bred as homozygotes in a C57Bl/6J background. All mouse experiments were approved by the Institutional Animal Care and Use Committee of the Agency for Science, Technology and Research (A*STAR; 231777) and performed in compliance with all of the relevant ethical regulations. All animals were placed under a 12 h light/12 h dark cycle. The room temperature was maintained at 21 ± 1 °C with 55–70% humidity. All mice were bred and maintained under specific-pathogen-free conditions in individually ventilated cages. The maximum tumour size allowed by the Institutional Animal Care and Use Committee is 15 mm in any dimension and none of the experiments exceeded this limit. For all of the experiments, adult mice (not selected for sex, except for the FACS experiments) of a minimum age of 6–8 weeks were used.
Tamoxifen induction
Adult mice aged 6–12 weeks were induced with a single or double intraperitoneal injection of 3.3–133 mg tamoxifen per kg bodyweight (Sigma–Aldrich) dissolved in corn oil. Double and triple injections were performed at 2-d intervals.
Tissue collection
Mice were euthanized by carbon dioxide asphyxiation before tissue collection. The peritoneal cavity was opened with scissors and the connective tissue was removed from the organs using blunt forceps. The small intestine and colon were flushed with cold Hanks’ Balanced Salt Solution (HBSS; Gibco). All of the organs were cut open longitudinally and the remaining faeces were removed by washing in HBSS.
Tissue fixation
Organs were incubated overnight at 4 °C (for immunochemistry or immunofluorescence) or 24 h at room temperature (for RNAscope) in 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS) (wt/vol). For tissue processing in the RNAscope experiments, all tools were carefully cleaned with RNaseZap (Invitrogen) to prevent RNA degradation.
Crypt isolation and cell sorting
Mouse small intestine, colorectal or caecum tissues were incubated in chelation buffer (5.6 mM sodium phosphate, 8 mM potassium phosphate, 96.2 mM sodium chloride, 1.6 mM potassium chloride, 43.4 mM sucrose and 54.9 mM d-sorbitol in Milli-Q water) with 10 mM (colorectum and caecum) or 2 mM (small intestine) ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol and 1 mM N-acetylcysteine for 2 h (colorectum and caecum) or 45 min (small intestine) at 4 °C on a roller. Crypts were isolated by repeated pipetting of finely chopped tissue in cold chelation buffer. Chelation buffer containing isolated crypts was filtered through a 100-μm filter mesh and centrifuged at 720g at 4 °C for 3 min. The pellet was resuspended in TrypLE (Life Technologies) with DNase I (0.8 U μl−1) (Sigma–Aldrich) and incubated at 37 °C for 10 min with intermittent trituration for digestion into single cells. Digestion was quenched by dilution with cold HBSS buffer. The suspension was centrifuged at 720g at 4 °C for 3 min. The pellet was resuspended in 2% foetal bovine serum (colorectum and caecum) or 20% foetal bovine serum + 5 mM EDTA and filtered through the 4-μm filter mesh cap of the FACS tube. Before sorting, 4′,6-diamidino-2-phenylindole (DAPI) was added at a final concentration of 1 μg ml−1 to stain dead cells. Single cells were sorted on a BD Influx Cell Sorter (BD Biosciences). Cells were collected in RLT Plus buffer (Qiagen) supplemented with β-mercaptoethanol (1/100) for RNA extraction, or advanced DMEM:F-12 medium (Gibco) supplemented with 0.5% Matrigel (Corning) for organoid culture.
RNA-seq and transcriptomic comparison
Single LGR5-GFPhigh and -GFPneg small intestinal or colonic epithelial cells from Lgr5-2A-eGFP mice were isolated by FACS (as described in the section ‘Crypt isolation and cell sorting’) and collected in RLT Plus + β-mercaptoethanol (1/100). A minimum of 30,000 cells for the colon and 50,000 cells for the small intestine were collected from each GFP fraction. Total RNA extracted from each fraction using the RNeasy Micro kit (Qiagen) was dissolved in RNase-free water and sent for sequencing at Novogene in Singapore. FASTQ files containing the RNA-seq reads were checked for quality control issues using FASTQC and MultiQC for which no issues were found. The reads were then mapped to the mouse genome build mm10 using STAR based on GENCODE M22 annotations. Gene counts were counted using featureCounts (part of the Subread package) based on GENCODE M22 annotations. The gene counts were then used in edgeR for a differential expression of genes analysis with multiple testing correction done using the Benjamini–Hochberg method.
Single-cell RNA-seq and analysis
10x Genomics scRNA-seq data were obtained as FASTQ files, which were processed with Cell Ranger version 7.1.0 using the mouse mm10 annotations for each individual tissue. For each tissue, the cell gene counts were imported into Seurat for independent data analysis. Cells were filtered to ensure that the number of detected genes was greater than 1,000 and the mitochondrial percentage was less than 20%. The top 2,000 variable features of the log-normalized counts data were used for principal component analysis. The top ten principal components (determined via an elbow plot) were then used to generate the SNN group, from which clusters were determined at a resolution of 0.5. Marker genes for each of the clusters were determined using the FindAllMarkers function in the Seurat package. These marker genes were then manually annotated to identify the various cell types. UMAP was performed on the top ten components to visualize the data. All analyses and visualizations were conducted in R version 4.2.1.
Organoid culture
FACS-isolated single cells were resuspended in growth factor-reduced Matrigel, seeded in 48-well plates and cultured in IntestiCult organoid growth medium (Stem Cell Technologies) supplemented with 20% Afamin/Wnt3a CM (MBL Life Science) and 100–200 μg ml−1 primocin (Invivogen), plus 10 μM Y-27632 (Sigma–Aldrich) for the first 2 d to prevent anoikis. Caecum organoid medium was supplemented with 100 ng ml−1 recombinant fibroblast growth factor 10 (PeproTech). The medium was refreshed every 2–3 d. Organoids were passaged when confluent, at least once a week. For RNA isolation, the well was washed with PBS before incubation in RLT Plus with β-mercaptoethanol (1/100). For differentiation assays, organoids were passaged and put in culture for 2 d in IntestiCult organoid growth medium and the medium was then changed to IntestiCult organoid differentiation medium (Stem Cell Technologies) supplemented with 2.5 µM DAPT (ab120633; Abcam) for 3 or 5 d for the distal colon and caecum, respectively.
IHC, immunofluorescence and RNAscope
IHC and immunofluorescence were performed following standard protocols. Briefly, fixed samples were dehydrated before embedding in paraffin blocks. Then, 7-μm sections were dewaxed and rehydrated before antigen retrieval was performed in a pressure cooker using a commercial solution at pH 9 (Dako). Samples were then incubated with hydrogen peroxide to inhibit endogenous peroxidase before being blocked for antibody incubation overnight at 4 °C. The primary antibodies used were rabbit anti-eGFP (2956S; Cell Signaling Technology; 1:200), goat anti-eGFP (Ab6673; Abcam; 1:500), rabbit anti-Ki-67 (MA5-14520; Thermo Fisher Scientific; 1:200), rabbit anti-RFP (600-401-379; Rockland Immunochemicals; 1:2,000), mouse anti-RFP (129244; Abcam; 1:500), mouse anti-β-catenin (610154; BD Transduction Laboratories; 1:250), mouse anti-E-cadherin (610181; BD Transduction Laboratories; 1:500), rabbit anti-Vimentin (ab92547; Abcam; 1:1,000), rabbit anti-α-SMA (MA5-11547; Invitrogen; 1:200), rabbit anti-ChgA (Ab15160; Abcam; 1:200), rabbit anti-Muc2 (PA5-21329; Thermo Fisher Scientific; 1:750), rabbit anti-Dra (NBP1-84450; Novus Biologicals; 1;500), rabbit anti-phospho-MAPK (4370S; Cell Signaling Technology; 1:200) and mouse anti-Trp53 (2524; Cell Signaling Technology; 1:200). For β-catenin IHC, 4-μm sections were collected and antigen retrieval was performed in Tris/EDTA pH 9.0 (ref. 10). On the following day, slides were washed with PBS and incubated with secondary antibodies. For IHC, the secondary antibody used was the peroxidase-conjugated anti-rabbit EnVision+ (Dako) and the staining was amplified by incubating the samples with 3,3′-diaminobenzidine (Dako). For immunofluorescence, the secondary antibodies used were goat anti-rabbit Alexa Fluor 488, goat anti-mouse-Alexa Fluor 647 and donkey anti-goat Alexa Fluor 647 (Invitrogen; 1:500). Samples were counterstained with haematoxylin (for IHC) or DAPI (for immunofluorescence) before mounting for imaging.
RNAscope was performed using a RNAscope 2.5 High-Definition Brown Assay kit (Advanced Cell Diagnostics) following the manufacturer’s recommendations. PPIB and DapB RNA probes were used as positive and negative controls, respectively.
Microscopy imaging and analysis
A Zeiss LSM 780 microscope was used to study the immunofluorescence slides. Brightfield images were captured on a Nikon NiE microscope with a DS-Ri2 camera, and stitching features were used. Images were saved as TIFF files and processed using Adobe Photoshop for figure mounting.
Quantitative PCR
FACS-sorted cells were lysed in RLT Plus buffer with β-mercaptoethanol before RNA extraction using an RNeasy Micro kit (Qiagen) according to the manufacturer’s instructions. Complementary DNA was generated using a SuperScript III kit (Life Technologies). qPCR was performed in triplicate per gene using SYBR green dye (Promega) according to the manufacturer’s instructions, and run on QuantStudio7 qPCR machines (Applied Biosystems). The data were analysed with the ∆∆Ct method using QuantStudio 7 software. Ct values were normalized using Gapdh housekeeping gene expression. The primer sequences are listed in Supplementary Table 7.
Statistics and reproducibility
For quantification of the traced crypts in the lineage tracing experiments, a minimum of 50 crypts per tissue region (colon proximal, middle, distal or rectum) were counted and the percentages of crypts retaining RFP expression were calculated. One representative section per mouse across three independent tracing experiments was selected for analysis. Mice with the correct genotypes were randomly assigned to control or treated groups. No animals or data points were excluded from the analyses. The data are presented as means ± s.e.m. The data distribution was assumed to be normal, but this was not formally tested. Statistical analyses were performed using GraphPad Prism (GraphPad software). Statistical significance was analysed by paired two-tailed t-test. P values are indicated in the relevant graphs. Each experiment was repeated at least three times independently. Data collection and analysis were not performed blind to the conditions of the experiments. Precise numbers of replicates are indicated in the figure captions. No statistical method was used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications20,32.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Whole colon and small intestine RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE303295. Two-part colon and three-part small intestine RNA-seq data that support the findings of this study have been deposited in the GEO under accession code GSE303433. Colon and small intestine region scRNA-seq data that support the findings of this study have been deposited in the GEO under accession code GSE303434. Previously published pylorus microarray data that were re-analysed here are available from the GEO under accession code GSE121803. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Tan, D. W. & Barker, N. Intestinal stem cells and their defining niche. Curr. Top. Dev. Biol. 107, 77–107 (2014).
Kiela, P. R. & Ghishan, F. K. Physiology of intestinal absorption and secretion. Best. Pract. Res. Clin. Gastroenterol. 30, 145–159 (2016).
Martinez-Guryn, K., Leone, V. & Chang, E. B. Regional diversity of the gastrointestinal microbiome. Cell Host Microbe 26, 314–324 (2019).
Hall, C., Youngs, D. & Keighley, M. R. Crypt cell production rates at various sites around the colon in Wistar rats and humans. Gut 33, 1528–1531 (1992).
Brown, K., Abbott, D. W., Uwiera, R. R. E. & Inglis, G. D. Removal of the cecum affects intestinal fermentation, enteric bacterial community structure, and acute colitis in mice. Gut Microbes 9, 218–235 (2018).
Den Besten, G. et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340 (2013).
Imperial, R. et al. Comparative proteogenomic analysis of right-sided colon cancer, left-sided colon cancer and rectal cancer reveals distinct mutational profiles. Mol. Cancer 17, 177 (2018).
Xi, Y. & Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 14, 101174 (2021).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Akyol, A. et al. Generating somatic mosaicism with a Cre recombinase–microsatellite sequence transgene. Nat. Methods 5, 231–233 (2008).
Boutin, A. T. et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev. 31, 370–382 (2017).
Kawaguchi, Y. et al. Mouse model of proximal colon-specific tumorigenesis driven by microsatellite instability-induced Cre-mediated inactivation of Apc and activation of Kras. J. Gastroenterol. 51, 447–457 (2016).
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017).
Tetteh, P. W. et al. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc. Natl Acad. Sci. USA 113, 11859–11864 (2016).
Xue, Y., Johnson, R., Desmet, M., Snyder, P. W. & Fleet, J. C. Generation of a transgenic mouse for colorectal cancer research with intestinal Cre expression limited to the large intestine. Mol. Cancer Res. 8, 1095–1104 (2010).
Seishima, R. et al. Neonatal Wnt-dependent Lgr5 positive stem cells are essential for uterine gland development. Nat. Commun. 10, 5378 (2019).
Van der Flier, L. G. et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136, 903–912 (2009).
Habowski, A. N. et al. Transcriptomic and proteomic signatures of stemness and differentiation in the colon crypt. Commun. Biol. 3, 453 (2020).
Tan, S. H. et al. AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature 578, 437–443 (2020).
Suh, Y. A. et al. Cell transformation by the superoxide-generating oxidase Mox1. Nature 401, 79–82 (1999).
Van der Post, S., Birchenough, G. M. H. & Held, J. M. NOX1-dependent redox signaling potentiates colonic stem cell proliferation to adapt to the intestinal microbiota by linking EGFR and TLR activation. Cell Rep. 35, 108949 (2021).
Hayashi, M. et al. Enteroendocrine cell lineages that differentially control feeding and gut motility. eLife 12, e78512 (2023).
Baker, D. et al. A cellular reference resource for the mouse urinary bladder. Preprint at bioRxiv https://doi.org/10.1101/2021.09.20.461121 (2021).
Munoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31, 3079–3091 (2012).
Azkanaz, M. et al. Retrograde movements determine effective stem cell numbers in the intestine. Nature 607, 548–554 (2022).
Corominas-Murtra, B. et al. Stem cell lineage survival as a noisy competition for niche access. Proc. Natl Acad. Sci. USA 117, 16969–16975 (2020).
Ritsma, L. et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365 (2014).
Zhan, T., Rindtorff, N. & Boutros, M. Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017).
Loree, J. M. et al. Classifying colorectal cancer by tumor location rather than sidedness highlights a continuum in mutation profiles and consensus molecular subtypes. Clin. Cancer Res. 24, 1062–1072 (2018).
Yamauchi, M. et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 61, 847–854 (2012).
Fatehullah, A. et al. A tumour-resident Lgr5+ stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat. Cell Biol. 23, 1299–1313 (2021).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Vogelstein, B. et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532 (1988).
Pedrazzini, T. et al. Cardiovascular response, feeding behavior and locomotor activity in mice lacking the NPY Y1 receptor. Nat. Med. 4, 722–726 (1998).
Gallois, C., Pernot, S., Zaanan, A. & Taieb, J. Colorectal cancer: why does side matter? Drugs 78, 789–798 (2018).
Hossain, M. S. et al. Colorectal cancer: a review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers (Basel) 14, 1732 (2022).
Xie, X. et al. Differences between carcinoma of the cecum and ascending colon: evidence based on clinical and embryological data. Int. J. Oncol. 53, 87–98 (2018).
Wong, C. C. & Yu, J. Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 20, 429–452 (2023).
Bras, M. M., Sousa, S. R., Carneiro, F., Radmacher, M. & Granja, P. L. Mechanobiology of colorectal cancer. Cancers (Basel) 14, 1945 (2022).
Xie, Y. et al. Impact of tumor site on lymph node status and survival in colon cancer. J. Cancer 10, 2376–2383 (2019).
Song, S., Wang, J., Zhou, H., Wang, W. & Kong, D. Poorer survival in patients with cecum cancer compared with sigmoid colon cancer. Med. (Kaunas) 59, 45 (2022).
Bailey, C. E. et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 150, 17–22 (2015).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Acknowledgements
We thank all members of N.B.’s group for technical expertise and insightful discussions; E. Wong for help with generating the mouse lines; S. Mustafah and staff at the A*STAR Singapore Immunology Network Flow Facility for assistance with FACS; staff at the A*STAR Advanced Molecular Pathology Laboratory for pathology evaluation; the A*STAR Microscopy Platform and its staff for assistance with confocal imaging; and E. Cheah and staff at the A*STAR Singapore Bioimaging Consortium–Nikon facility for assistance with brightfield imaging. This study is supported by the A*STAR Institute of Molecular and Cell Biology, National Research Foundation (under NRFI2017-03) and Open Fund Individual Research Grant (OFIRG24Jul-0078).
Author information
Authors and Affiliations
Contributions
M.G. designed and performed all of the empirical experiments, collected and analysed the data and wrote the manuscript. S.S., T.C.-Y.C. and S.Y. performed all of the empirical experiments and collected and analysed the data. T.C.-Y.C. performed the mouse husbandry. S.Y. cloned and generated the Nox1-2A-eGFP and Nox1-2A-CreERT2 mouse lines. S.Y., M.G. and T.C.-Y.C. cloned and generated the Npy1r-eGFP-IRES-CreERT2 mouse line. Y.Y. initiated the project and performed the RNA-seq experiment. S.P. and B.L. analysed the RNA-seq and scRNA-seq data and performed the transcriptomics comparison and gene expression analysis. H.Y.G.L. provided critical analyses and wrote the manuscript. N.B. designed and supervised the project, analysed the data and wrote the manuscript. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Louis Vermeulen, Omer Yilmaz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparative expression profiling of Lgr5+ stem cell populations along the GI tract identifies novel regional markers.
a) Venn diagram representing the overlap between the top 200 differentially expressed genes (DEG) of the Lgr5+ enriched transcriptome using the established Lgr5-EGFP-IRES-CreERT2 (5KI) mouse and an independent Lgr5-2A-EGFP reporter mouse; b) Only 9 genes were identified as potentially enriched in proximal colon stem cells. None of these genes could be validated; c) Graph representing the percentage of Npy1r+ cells within the enteroendocrine cluster for each tissue compartment; d) (Upper panel) Graph representing the percentage of Lgr5+ cells per cluster and their co-expression with Nox1, Npy1r, and both Nox1 and Npy1r; (Lower panel) Graph representing the total number of Lgr5+ cells per cluster; e) (Upper panel) Graph representing the percentage of Nox1+ cells per cluster and their co-expression with Lgr5, Npy1r, and both Lgr5 and Npy1r; (Lower panel) Graph representing the total number of Nox1+ cells per cluster; f) (Upper panel) Graph representing the percentage of Npy1r+ cells per cluster and their co-expression with Lgr5, Nox1, and both Lgr5 and Nox1; (Lower panel) Graph representing the total number of Npy1r+ cells per cluster.
Extended Data Fig. 2 Nox1 and Npy1r are not enriched in small intestine stem cell clusters.
a) Heatmap representing the level of expression of identified key lineage markers for each cluster in the proximal, middle, and distal small intestine; b) UMAP plots representing all the clusters identified (left panel) before being reassigned in different lineage clusters using the expression heatmap in a) (right panel); c) UMAP plots representing the distribution of Lgr5+ (upper panel), Nox1+ (middle panel), and Npy1r+ cells (lower panel) for each intestinal region (proximal, middle, and distal small intestine). Stem cell (SC) cluster is delineated in blue dashed line and enteroendocrine (EE) cluster is delineated in brown dashed line according to the lineage clusters defined in b). Assay was performed on one biological sample per tissue region.
Extended Data Fig. 3 Restricted expression of Nox1 and Npy1r in stem cell/early TA compartments of the Ileum and Peyer’s patches of the SI, and in stromal cells of the bladder respectively.
a) RNAscope analysis for Nox1 showed no expression in the stomach (corpus and pylorus) and the proximal part of the small intestine (duodenum and jejunum) (n = 5); b) RNAscope analysis for Nox1 and Lgr5 showed that Nox1 was expressed in the early transit-amplifying compartment and lowly expressed in the CBC cells of the ileum. Stronger Nox1 expression primarily in the CBC compartment was detected in intestinal crypts directly adjacent to the Peyer’s patches (n = 5). Green arrows indicate the +4 cell position while red stars indicate the CBC cell position; c) RNAscope analysis for Npy1r showed no expression in the stomach (corpus and pylorus) and the small intestine epithelium (n = 5); d) RNAscope analysis of Npy1r showed a subset of stromal cells expressing Npy1r in the bladder stroma (n = 5). Scale bar: 25μm.
Extended Data Fig. 4 Nox1- and Npy1r-GFP reporters recapitulate the endogenous expression seen in the small intestine and bladder.
a) GFP IHC performed on Nox1-2A-EGFP mouse line was observed in the +4 to +6 presumptive TA cells and the CBC cells in the ileum at a lower level. This expression was detected at stronger level in the same cells of the crypt bordering the peyer’s patch. Green arrows indicate the +4 cell position while red stars indicate the CBC cell position (n = 5); b) Co-IF of GFP with the epithelial marker E-cadherin, the mesenchyme marker Vimentin, and the muscle marker a-SMA showed that GFP+ cells were Vimentin+ and excluded from the urothelium and muscle cells (n = 5). Scale bar: 25μm (a) and 50μm (b).
Extended Data Fig. 5 Nox1+ colon cells express markers of undifferentiated stem cells.
a) qPCR analysis on Nox1- and Npy1r-GFP FAC-sorted cells showed an enrichment in stem cell markers Lgr5 (Nox1 caecum n = 3, 12.42 ± 1.219; Nox1 colon n = 4, 14.0825 ± 1.656; Npy1r colon n = 6, 9.6516 ± 0.5590), Smoc2 (Nox1 caecum n = 3, 5.8765 ± 0.2463; Nox1 colon n = 4, 7.5078 ± 0.3144; Npy1r colon n = 4, 6.368 ± 0.7178), Ascl2 (Nox1 caecum n = 3, 7.613 ± 1.399; Nox1 colon n = 4, 17.916 ± 0.7981; Npy1r colon n = 4, 11.0515 ± 2.212) (upper panel). Lineage markers Dra (Nox1 caecum n = 4, 0.079 ± 0.03201; Nox1 colon n = 5, 0.18 ± 0.04231; Npy1r colon n = 7, 0.19 ± 0.03690), Muc2 (Nox1 caecum n = 3, 0.2367 ± 0.09939; Nox1 colon n = 3, 0.18 ± 0.06245; Npy1r colon n = 7, 0.09 ± 0.01952), ChgA (Nox1 caecum n = 3, 0.1667 ± 0.1217; Nox1 colon n = 3, 0.12 ± 0.04041; Npy1r colon n = 7, 0.7814 ± 0.1142), and Reg4 (Nox1 caecum n = 3, 0.15 ± 0.04807; Nox1 colon n = 3, 0.3967 ± 0.1530; Npy1r colon n = 7, 0.57 ± 0.1214) were upregulated in the GFP− population, indicating that Nox1+ and Npy1r+ cells enrich for stem cells. Statistical significance was determined by two-tailed t-test; b-c) Co-IF for GFP and lineage markers using Nox1-2A-EGFP (n = 5) (b) and Npy1r-EGFP-IRES-CreERT2 (n = 5) (c) mice colon (caecum, distal colon and rectum) showed that GFP+ cells do not overlap with the lineage markers. Co-IF with Ki67 indicated that Nox1+ and Npy1r+ cells are proliferative. Scale bar: 25μm.
Extended Data Fig. 6 Nox1-2A-CreERT2 induction is highly efficient in the caecum.
a) Graph representing the percentage of crypts retaining the RFP label for each tissue compartment (Caecum: 97.77% ± 0.49%; CM: 91.71% ± 4.86%; CD: 92.53% ± 4.23%; Rectum: 68.54% ± 5.31%; n = 3) at one month post-induction of adult Nox1-2A-CreERT2, LSL-TdTomato mice; b) Nox1-2A-CreERT2, TdTom mice were injected with 0.1 mg of tamoxifen per 30 g of body weight. Colon compartments were harvested at 24 h, 1-week and 1-month post-induction. At 24 h, all the cells at the base of the caecum crypts were RFP+ while only 1 cell on average was labelled within the middle and distal colon. Insets represent a higher magnification of a representative crypt; After 1-week and 1-month, RFP+ crypt frequency significantly decreased in the middle and distal colon while the RFP+ crypt rate remained constant over time in the caecum (n = 3 mice each); c) Quantification of the percentage of RFP+ crypts following 24 h (Caecum n = 10, 70.40% ± 3.47%; Middle colon n = 6, 95.67% ± 1.41%; Distal colon n = 6, 87.33% ± 2.51%), 1 week (Caecum, 58.44% ± 4.75%; Middle colon, 48.44% ± 4.79%; Distal colon, 44.00% ± 2.96%; n = 9) and 1 month (Caecum, 61.33% ± 3.00%; Middle colon, 31.11% ± 3.04%; Distal colon, 34.44% ± 2.47%; n = 9) tracing. Statistical significance was determined by two-tailed t-test. Scale bar: 25μm.
Extended Data Fig. 7 A subset of Nox1-expressing cells in the small intestine is self-renewing and multipotent.
a) Nox1-2A-CreERT2, Rosa26-LSL-tdTomato mice were injected with 3 mg/30 g body weight of tamoxifen and harvested after 24 h, 1-week, and 1-month. At 24 h, majority of the cells in the vicinity of the +4 cell were RFP +. Only a small subset of CBC cells were RFP + . After one month of tracing, the crypts and villi in the ileum were partially covered by RFP-expressing cells. Red asterisks represent RFP + CBC cells; b) Frequency of RFP+ cells increased in the crypts adjacent to the Peyer’s patches after 24 h induction. All cells within the crypt-villus units adjacent to the Peyer’s patches were RFP+ at 1-month. Scale bar: 25μm.
Extended Data Fig. 8 Npy1r is expressed in long-lived stromal cells of the bladder.
a) Graph representing the percentage of crypts retaining the RFP label for each tissue compartment (CM: 22.67% ± 3.53%; CD: 74.79% ± 4.40%; Rectum: 81.79% ± 4.04%; n = 3) at one month post-induction in adult Npy1r-EGFP-ires-CreERT2, LSL-TdTomato mice; b) Npy1r- EGFP-ires-CreERT2, Rosa26-LSL-tdTomato mice were injected with 4 mg/30 g body weight of tamoxifen and harvested after 24 h (n = 3), 1-week (n = 3), and 1-month (n = 3). At 24 h, RFP was detected in stromal cells. This pool of cells was maintained 1 week and 1 month after induction; c) Co-IF of RFP with the epithelial marker E-cadherin, the mesenchyme marker Vimentin, and the muscle marker a-SMA showed that RFP+ cells were Vimentin+ and excluded from the urothelium and muscle cells (n = 3). Scale bar: 50μm.
Extended Data Fig. 9 Nox1 CreERT2-driven Apc deletion in the intestine generates mild hyperplasia only in crypts adjacent to Peyer’s patches.
a) Nox1-2A-CreERT2, APCfl/fl 5-month old mice were harvested and stained for RFP and β-catenin in all the digestive tissues expressing Nox1 to evaluate any potential Cre recombinase expression leakage (n = 3); b) Mice were injected twice at 2-days interval with 4 mg/30 g body weight of tamoxifen and the small intestine harvested after 1 month. β-catenin IHC showed nucleocytoplasmic accumulation only in the crypts directly adjacent to Peyer’s patches but absent from the ileum (n = 4); c) Co-IF of β-catenin with Ki67 showed that these hyperplastic crypts were proliferative (n = 4); d) Nox1-2A-CreERT2, APCfl/fl mice were bred with LSL- KrasG12D mice. Mice were induced with 1 mg of tamoxifen per 30 g of body weight and harvested after 1 month. No β-catenin accumulation was detected in the ileum and Peyer’s patch epithelium and hyperactivation of pMapk was found as expected in the CBC and TA cells (n = 3). Scale bar: 250μm (a, upper panel), 50μm (a, lower panel), and 25μm (b-d).
Extended Data Fig. 10 Npy1r CreERT2-driven Apc deletion does not initiate tumour formation in the bladder.
a) 3-month old Npy1r-EGFP-IRES-CreERT2, APCfl/fl mice were harvested and stained for RFP in the entire colorectum to evaluate any potential Cre recombinase expression leakage. No RFP staining was detected (n = 3); b) Npy1r-EGFP-IRES-CreERT2, APCfl/fl mice were injected with 3 doses of 4 mg tamoxifen per 30 g body weight every 2 days and harvested 8 weeks post- induction. No β-catenin accumulation was detected in the bladder (n = 6). Scale bar: 500 μm.
Supplementary information
Source data
Source Data Fig. 4
Statistical source data and qPCR data.
Source Data Extended Data Fig. 5
qPCR data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gasnier, M., Chen, T.CY., Yada, S. et al. NOX1 and NPY1R mark regional colon stem cell populations that serve as cancer origins in vivo. Nat Cell Biol 27, 1632–1646 (2025). https://doi.org/10.1038/s41556-025-01763-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01763-1
This article is cited by
-
Stem cell sources of colon cancer in mice
Nature Reviews Molecular Cell Biology (2025)