Abstract
Epigenetic modifications on natural chromosomes are inherited and maintained in a default state, making it challenging to remove intrinsic marks to study the fundamental principles of their establishment and further influence on transcriptional regulation. In this study, we developed SynNICE, a method for assembling and delivering intact, naive, synthetic megabase (Mb)-scale human DNA into early mouse embryos, to study de novo epigenetic regulation. By assembling and delivering a 1.14-Mb human AZFa (hAZFa) locus, we observed the spontaneous incorporation of murine histones and the establishment of DNA methylation at the one-cell stage. Notably, DNA methylation from scratch strongly enriches at repeat sequences without H3K9me3 reinforcement. Furthermore, the transcription of hAZFa initiated at the four-cell stage is regulated by newly established DNA methylation. This method provides a unique platform for exploring de novo epigenomic regulation mechanisms in higher animals.
Similar content being viewed by others
Main
The process of genome synthesis has facilitated the construction of viral genomes1, bacterial genomes2,3 and eukaryotic chromosomes4,5,6, paving the way for genome writing in mammalian systems7. Currently, mammalian genome rewriting efforts have focused primarily on individual genes with non-coding regulatory elements, approximately 100 kilobases (kb) in length, by current synthetic DNA methods8,9,10. Functional reconstitution by mammalian genome rewriting at the Mb-level capacity stays demanding, which is crucial for a comprehensive exploration of spatiotemporal gene expression patterns, encompassing both coding and non-coding regulatory genome sequences at a multi-gene level11. However, how the cellular environment remodels and regulates a synthetic, naive, Mb-scale human DNA remains challenging. Furthermore, dissecting de novo epigenetic modifications at temporal resolutions can provide a comprehensive understanding of mammalian epigenetic and transcriptional regulation from a bottom-up perspective.
In mammals, epigenomic modifications exhibit different patterns in various cells in a cell-lineage-dependent manner12. The totipotent mouse early embryo exhibits well-elucidated mechanisms underlying epigenetic reprogramming and zygotic genome activation13,14, making it suitable for studying de novo epigenomic modification processes and transcriptional regulation on synthetic, naive, Mb-scale DNA at temporal resolution. However, examining the remodeling process at a temporal resolution still requires addressing two key challenges: (1) the feasible synthesis and assembly of Mb-scale DNA with designer sequences corresponding to potentially any human genome locus, including conserved highly repetitive sequences, and (2) precisely and efficiently delivering such large, intact DNA molecules into totipotent mammalian cells15. Developing robust and efficient DNA delivery methods capable of transferring Mb-scale constructs across diverse organisms remains a critical and urgent challenge in the field. Although Escherichia coli is effective for Mb-scale DNA assembly16, from preexisting bacterial artificial chromosomes (BACs), the system cannot stably maintain DNA fragments larger than 500 kb (ref. 17). Naked, large DNA molecules extracted from E. coli contain interfering bacterial methylation18. Obtaining high-concentration and high-purity large DNA constructs is extremely challenging, and they are also highly susceptible to physical breakage in vitro, resulting in very low efficiency19. In addition, transfection or electroporation of naked DNA leaves out the natural chromatin structure, making it insusceptible to chromatin-based regulatory mechanisms within the cellular environment. An improved strategy using the DNA assembly host as a cellular transfer shuttle can avoid frequent breakage during extraction and purification processes.
Yeast enables Mb-scale DNA assembly through stepwise SwAP-In and hierarchical method20,21,22,23, but multi-fragment recombination of highly repetitive sequences leads to lower success rates24. To address this, a combinatorial assembly strategy is needed to reduce the mismatch probability and achieve rapid, precise de novo assembly of chemically synthesized, highly repetitive, human Mb-scale DNA. Delivering large DNA to cultured mammalian cell lines through PEG-mediated cell fusion with yeast protoplasts was previously reported25,26, but its low efficiency and long-term selection restricted the ability to study de novo epigenetic regulation with temporal resolution. The ortholog proteins and molecules present in the yeast cytoplasm27 also influence early embryo development and decrease delivery efficiency. Thus, a more precise and efficient Mb-scale DNA transfer shuttle would be valuable for mammalian genome synthesis.
To fill in these gaps, we developed the SynNICE method to precisely assemble and directly deliver an intact, naive, Mb-scale synthetic DNA into early mouse embryos, enabling the study of de novo epigenetic regulation on human gene sequence. Our method involves chemically synthesizing and assembling Mb-scale human genome regions in yeast, followed by the Nucleus Isolation for Chromosomes Extraction (NICE) technique to isolate yeast nuclei with intact chromosomes. To validate our method, we designed a 1.14-Mb DNA according to the hAZFa locus, a single-copy segment located in the human Y chromosome that appeared in evolution after the divergence between humans and chimpanzees28,29 and lacked gene dosage effects and homologous interference when introduced into mice. We delivered hAZFa into mouse parthenogenetic embryos and further explore cross-species remodeling processes and transcriptional regulation at a temporal resolution (Fig. 1).
Results
De novo assembly of Mb-scale highly repetitive human DNA
In this study, we focused on the AZFa sequence as microdeletion of AZFa leads to a severe form of male infertility without any available clinical solution30. We sequenced two infertile patients with AZFa microdeletion and found that both had lost a subregion of approximately 798 kb between two long homologous human endogenous retroviral sequences (HERV15yq1/HERV15yq2) (Extended Data Fig. 1 and Supplementary Table 1). We chose NCBI36/hg18 as the reference genome (chrY: 12,952,206–14,101,944 base pairs (bp)), and two HERV15 blocks were deleted (Fig. 2a). We inserted 12 28-bp watermark sequences into the introns of both coding and non-coding genes (Supplementary Table 2). Notably, the hAZFa region exhibited a significantly higher proportion of repetitive sequences at 69.38% compared to the genomes of Saccharomyces cerevisiae and E. coli (Fig. 2b and Extended Data Figs. 2a–c).
a, The AZFa locus and UCSC database gene annotations are indicated. The hAZFa was designed to split into four large fragments—SynA, SynG, SynB and SynC—with two HERV15 block deletions. b, The vertical blue lines indicate the position of repetitive elements, including STRs, LTRs, LINEs and SINEs. c, Circos plot showing the assembly of hAZFa. Twelve 28-bp watermark sequences were inserted into the intron region of hAZFa. WM(n), watermark(n). d, The recombination-based combinatorial assembly strategy for Mb-scale synthetic human DNA with highly repetitive sequences. e, Restriction digestion analysis of hAZFa followed by PFGE. Yeast_hAZFa: S. cerevisiae VL6-48 containing hAZFa; Yeast_Control: S. cerevisiae VL6-48 harboring pRS416 as a control. Left, AscI enzyme digestion, with VL6-48 genome as a marker. Right, BsiWI enzyme digestion, with λ PFG ladder as a marker. n = 3 biological replicates. f, Growth assay of Yeast_hAZFa compared to Yeast_Control. g, Transcriptome analysis of Yeast_hAZFa compared to Yeast_Control. A two-sided adjusted P value (q value) threshold of 0.001 (Wald test with Benjamini–Hochberg correction) and a |log2(FC)| > 1.0 were used. h, The length of isoforms on hAZFa compared to yeast endogenous genome. i, Proteomic analysis of Yeast_hAZFa compared to Yeast_Control. Differentially expressed genes were identified using a two-sided P value threshold of 0.001 (Welch’s t-test) with FC > 1.2 or FC < 0.83. j, Genome-wide Hi-C contact map of the Yeast_hAZFa chromosome at a 10-kb resolution (left). The Hi-C interaction heatmap of hAZFa is magnified on the right. FC, fold change; LINE, long interspersed nuclear element; LTR, long terminal repeat; SINE, short interspersed nuclear element; STR, short terminal repeat; PFG, pulsed-field gel.
Afterwards, we split this 1.14-Mb hAZFa into 233 of the 5.5-kb DNA fragments and chemically synthesized them through a commercial order (GenScript/GENEWIZ) (Fig. 2c and Supplementary Table 10). To assemble the Mb-scale highly repetitive human DNA sequence from these DNA fragments, we developed a combinatorial assembly strategy (Fig. 2d). This approach aimed to avoid the simultaneous assembly of very large fragments, instead focusing on combining multiple short DNA fragments with longer homologous arms (500 bp) for higher efficiency and accuracy. In the first step, we assembled 233 fragments into 23 larger segments ranging from 40 kb to 71 kb, achieving a nearly 10-fold reduction in fragment number. This was accomplished using chemical transformation and homologous recombination in S. cerevisiae BY4741 (Supplementary Table 3). The success rates varied from 1/108 to 33/48, except for three 55-kb fragments (A2, A5 and A6), which needed an additional assembly step. We divided them into 25-kb and 30-kb segments before reassembling them to create the desired 55-kb constructs. In the second step, we used protoplast transformation with two strains, S. cerevisiae VL6-48α and VL6-48a, with opposite mating types, to assemble the 23 fragments into four large constructs—SynA, SynG, SynB and SynC—ranging from 268 kb to 331 kb (Extended Data Fig. 3a). Although each of these four constructs was assembled from an equal number of six large fragments, the efficiency of SynA (331 kb) was lower than that of SynB (276 kb) and SynC (268 kb), indicating that assembly efficiency for repetitive sequences is affected by fragment length (Supplementary Table 4). In the third step, we incorporated yeast mating with CRISPR to parallel assemble Mb-scale constructs in two rounds. The MATα yeast containing a Cas9-only plasmid and SynA was crossed with the MATa yeast carrying a single guide RNA (sgRNA) expression plasmid and SynG. After yeast mating, the Cas9 and sgRNA were combined to cleave SynA and linearize SynG, allowing the SynA fragment to assemble into SynG by homologous recombination. SynBC was assembled similarly, with an efficiency of 90% for SynAG and 92% for SynBC (Extended Data Fig. 3b and Supplementary Table 5). After sporulation and another round of yeast mating, pulsed-field gel electrophoresis (PFGE) and deep sequencing analysis validated the successful assembly of intact 1.14-Mb hAZFa in yeast (Fig. 2e and Extended Data Fig. 3c). Taken together, we chemically synthesized and assembled a 1.14-Mb hAZFa region in yeast.
Mb-scale human DNA exhibits higher-order chromatin structure in yeast
We observed no significant growth difference between yeast containing the Mb-scale synthetic human AZFa (Yeast_hAZFa) and yeast containing control plasmid (Yeast_Control) (Fig. 2f). We performed RNA sequencing (RNA-seq) and found that only 2% (121/5,997) of endogenous genes had altered expression in Yeast_hAZFa (Fig. 2g and Supplementary Table 6). Although many transcription factors in human cells have functional orthologs in yeast31, transcriptome profiling indicated that no human annotated transcripts of hAZFa were expressed in yeast (Extended Data Fig. 4a). However, high levels of transcriptional noise occurred in yeast32, and we found that the average length of the hAZFa isoforms was approximately 0.77 kb (ranging from 54 bp to 3,691 bp), which was shorter than that of the yeast endogenous chromosome isoforms (~1.22 kb) (Fig. 2h). Proteomics was also performed on Yeast_hAZFa and Yeast_Control, revealing that only 1% (31/3,129) of endogenous genes had altered expression at the protein level (Fig. 2i). Kyoto Encyclopedia of Genes and Genomes pathway enrichment revealed an increase in lysine biosynthesis proteins and proteins associated with amino acid metabolism (Extended Data Fig. 4b), which correlates well with our transcriptomic data. Total RNA and protein yields from whole-cell extracts of Yeast_hAZFa were similar to those of Yeast_Control (Extended Data Fig. 4c,d). Overall, neither transcriptome nor proteome analysis indicated a strong differential regulation of the additional hAZFa in yeast.
The contact map from Hi-C assays demonstrated that hAZFa in yeast exhibited only strong self-interactions, highlighting the remarkable disparities in DNA sequences between humans and yeast (Fig. 2j). However, hAZFa still had limited centromere–centromere interactions with endogenous chromosomes (Extended Data Fig. 4e) due to its backbone vector containing yeast centromere sequences. The z-score difference contact map showed that the interactions among the 16 endogenous chromosomes of Yeast_hAZFa were much stronger on centromeres compared to those in the Yeast_Control strain (Extended Data Fig. 4f). We further calculated the ratio of the interaction frequency within the centromere region to the average interaction frequency between the centromere region and all regions of the chromosome at 10-kb resolution and defined it as relative interaction frequency (Supplementary Tables 7 and 8). The relative interaction frequency significantly increased the interactions between the centromeres of the endogenous chromosomes in the Yeast_hAZFa strain (Extended Data Fig. 4g). Taken together, our data indicate that the presence of large exogenous hAZFa within the host has limited influence on the transcriptome, proteome and three-dimensional (3D) structure.
Isolation of yeast nuclei containing intact Mb-scale human DNA
To transfer 1.14-Mb hAZFa from yeast directly into recipient cells, we purposed to use the yeast nucleus to protect synthetic DNA with the elimination of excess cellular components and substances. We developed NICE to isolate yeast nuclei while preventing nuclear DNA degradation and preserving chromosome structure without the use of crosslinking (Fig. 3a). Although previous studies successfully preserved a structurally intact nuclear envelope through spheroplasting, homogenization and purification33,34, PFGE results revealed complete degradation of endogenous chromosomal DNA (Fig. 3b, Lane 1). We speculated that mechanical disassembly of the cell membrane by homogenization inevitably activates endogenous DNase and leads to DNA degradation in nuclei35. Therefore, to reduce endogenous DNase activity, we added DNase inhibitor along with EDTA, which binds calcium and magnesium ions to prevent activation of Ca2+/Mg2+-dependent endonucleases throughout the lysis and purification process. We obtained some weak chromosome DNA bands, indicating the successful isolation of longer DNA fragments. However, the low yield indicated that significant degradation still occurred (Fig. 3b, Lane 2). We further attempted to preserve chromosomes in a compact state within isolated nuclei by collecting cells in the stationary phase and adding polyamines36 as crowding reagents (Fig. 3b, Lane 3). Indeed, by combining DNase inhibition and maintenance of chromosomal DNA condensation, we successfully isolated yeast nuclei containing intact endogenous chromosomes with minimal degradation (Fig. 3b, Lane 4). The presence of intact hAZFa in the isolated yeast nuclei was further confirmed by restriction enzyme digestion (Fig. 3b and Extended Data Fig. 5a). We performed the southern blotting following the PFGE analysis and revealed that 87.76% of hAZFa molecules were intact in isolated nuclei compared to Yeast_hAZFa (Fig. 3c and Extended Data Fig. 5b). PFGE results confirmed that chromosomal DNA remained intact after storage for over 6 months at −80 °C (Extended Data Fig. 5c). The nuclei isolated using our technology still maintained structural integrity with the nucleus size reduced to approximately 1 µm in sucrose solution (Fig. 3d). Taken together, our NICE approach for isolating yeast nuclei successfully protects the synthetic Mb-scale DNA from degradation in vitro.
a, Graphic depiction of yeast nuclei isolation with intact synthetic Mb-scale DNA, maintaining chromosome structure. b, PFGE analysis of the isolated yeast nuclei by NICE. Left, PFGE analysis of endogenous chromosomes. In Lane 1, nuclei were isolated according to the previous protocol; Lane 2 was optimized by adding DNase inhibitor; Lane 3 was optimized by adding spermine and spermidine; and Lane 4 was treated by adding both DNase inhibitor and spermine and spermidine. Analysis of hAZFa in isolated nuclei was performed by PFGE with AscI enzyme digestion, using VL6-48 genome as a marker (n = 3 biological replicates). c, Southern blot following PFGE analysis showing the integrity of full-sized hAZFa in isolated nuclei prepared by NICE, compared to in Yeast_hAZFa (n = 4 biological replicates). d, Staining for the isolated nucleus. DAPI staining for DNA and DiIC12(3) staining for yeast nuclear envelope. An overlay of the two signals (merge) is also shown. One representative image from three independent experiments is shown. Scale bars, 1 µm. e, The average signals of ATAC–seq enrichment around the central peak of hAZFa in isolated nuclei and yeast. Control: S. cerevisiae VL6-48 harboring pRS416. f, The average signals of H3K4me3 ChIP–seq enrichment around the peak center of hAZFa in isolated nuclei and yeast. g, Genome-wide Hi-C contact map of chromosomes in isolated nuclei at a 10-kb resolution (left). Zoom-in of the Hi-C interaction heatmap of hAZFa in isolated nuclei (right).
We further conducted several analyses to examine whether the hAZFa in isolated nuclei was still compacted into a chromosome. We first performed assay for transposase-accessible chromatin with sequencing (ATAC–seq) in both isolated nuclei and yeast. The ATAC–seq signals around the central peak of hAZFa were stronger in isolated nuclei than in yeast (Fig. 3e), and a large fraction of ATAC–seq peaks overlapped between the isolated nuclei and yeast (Extended Data Fig. 5d), suggesting that hAZFa in isolated nuclei still maintained chromatin accessibility and was protected by yeast nucleosome. Transposase-accessible chromatin with virtualization (ATAC–see) was performed on the isolated nuclei to test the heterogeneity of individual nuclei37. We observed that the signals of isolated nuclei were relatively uniform (Extended Data Fig. 5e). We also profile chromatin activity using histone H3K4me3 chromatin immunoprecipitation followed by sequencing (ChIP–seq) in both isolated nuclei and yeast. Signals around the peak center of hAZFa in isolated yeast nuclei were weaker than those in yeast (Fig. 3f and Extended Data Fig. 5f), suggesting that hAZFa in isolated nuclei still has an active epigenetic state with yeast H3K4me3 modification. Yeast immunostaining also indicated weaker H3K4me3 signals and less heterogeneity of isolated nuclei than in Yeast_hAZFa (Extended Data Fig. 5g). We further performed Hi-C to investigate the chromatin organization of hAZFa in isolated yeast nuclei. The contact map and 3D conformation showed that chromosomal DNA in the isolated nuclei maintained a 3D structure in spatial position (Fig. 3g and Extended Data Fig. 6a). The z-score difference contact map (Extended Data Fig. 6b) and relative interaction frequency (Extended Data Fig. 6c) indicated that interactions of endogenous chromosome centromeres were significantly weakened in isolated nuclei yeast compared to Yeast_hAZFa. In summary, these data confirm that the intact synthetic DNA at the Mb scale maintained its chromatin structure in vitro and was still wrapped by yeast nucleosomes.
Delivery of intact synthetic Mb-scale DNA into mouse embryos
After successfully obtaining yeast nuclei with intact hAZFa in vitro, we proceeded to deliver the nuclei into mouse metaphase II (MII) oocytes through nuclear microinjection. After that, we analyzed the remodeling processes during the parthenogenetic embryo development (Fig. 4a). The small yeast nuclei were visible under optical microscopy, and, on average, 2–4 yeast nuclei were injected into one oocyte. To confirm the successful delivery of the nucleus into mouse oocytes, we initially added lipophilic carbocyanine dye DiIC12(3) into yeast nuclei to target the nuclear membrane. After the injection, the oocyte was fixed and subsequently stained with 4′,6-diamidino-2-phenylindole (DAPI) for DNA, ensuring that the nucleus was properly delivered into the cytoplasm (Fig. 4b).
a, Graphic illustration of hAZFa remodeling in mouse early embryos. b, Staining for the isolated yeast nucleus after injection into mouse MII oocytes with immediate fixation. Yeast nuclei membrane stained with DiIC12(3) (red) and DNA stained with DAPI (blue). The dashed circle represents the location of murine chromosome DNA. Scale bars, 20 µm. A magnified view shows the yeast nucleus in the oocyte cytoplasm. Scale bars, 1 µm. Quantification of three replicate experiments of yeast nuclei injected into mouse MII oocytes. Sample sizes are as follows: n = 11, 11 and 11. c, Live images of murine histones H3.3 (green) and H2B (red) incorporated into the yeast nucleus at 6 hours after injection. Scale bars, 20 µm. A magnified view shows the yeast nucleus in the cytoplasm. Scale bars, 1 µm. Quantification of three replicate experiments of murine histone incorporation. Sample sizes are as follows: n = 16, 21 and 11. d, Still images of timelapse imaging of H3.3–GFP in mouse MII oocytes. Timepoints represent hours after injection of yeast nuclei. Yeast nuclei with H3.3–GFP incorporation are indicated by yellow dashed circles. h, hours; TSS, transcription start site.
Because yeast and murine histones are highly conserved38, we initially sought to determine if the murine histones in mouse early embryos could be spontaneously incorporated into yeast nuclear DNA. Initially, we selected H3.3, a conserved H3 variant, as the exclusive form of H3 present in S. cerevisiae39, and it plays a role in replication-independent nucleosome assembly40. We expressed green fluorescent protein (GFP)-tagged mouse H3.3 and conducted live-cell imaging to observe the deposition. We observed the incorporation of mouse H3.3 into the yeast nucleus in the cytoplasm and quantified the intensity of signals (Fig. 4c). We also identified the presence of mCherry-tagged mouse H2B, which requires interaction with the H3–H4 tetramer for incorporation, and we discovered that mouse H2B was incorporated simultaneously (Fig. 4c). We wondered whether the spontaneous incorporation relied on parthenogenetic activation, and we found that the H3.3 signals appeared very soon in mouse oocytes (Fig. 4d). The results indicated that the chromatin dynamics were not dependent on the fertilization process. In addition, we also performed CUT&Tag to observe the histone mark H3K9me3 on hAZFa in early embryos, which is absent from endogenous S. cerevisiae41. The H3K9me3 signals of hAZFa were rare (Extended Data Fig. 7a), and distribution did not form enriched domains, indicating that H3K9me3 was not well established in that region. We also compared the H3K9me3 distribution of mouse genome regions in injected hAZFa embryos with previous H3K9me3 ChIP–seq data of mouse wild-type embryos and confirmed that our quality chromatin data recapitulate results of endogenous H3K9me3 pattern (Extended Data Fig. 7b). These findings suggested that nucleosome replacement and histone modification rapidly occurred on the endogenous chromosomal DNA upon entry into the cells.
De novo DNA methylation of synthetic human DNA
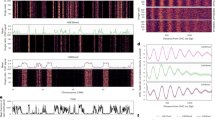
Because S. cerevisiae naturally lacks cytosine methylation42, we, therefore, wanted to know whether DNA methylation was also initiated in hAZFa. De novo DNA methylation and hydroxymethylation of the yeast nucleus are quickly established in mouse early embryo at 8 hours after injection, as revealed by staining for 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) as markers (Fig. 5a). We then performed whole-genome bisulfite sequencing (WGBS) to map the de novo DNA methylation profile of hAZFa compared to the endogenous AZFa region in human sperm and zygote43. The biological replicates showed highly reproducible results of de novo DNA methylation that occurred on hAZFa (Fig. 5b). In parallel to non-methylated hAZFa DNA in yeast, the methylation level of entire hAZFa in early parthenogenetic embryos was 9.9% (Extended Data Fig. 7c). The hAZFa contains 8,990 CpG sites, and 99% of these have been covered. We found that 46.35% of the sites were methylated in mouse one-cell embryos (Fig. 5c). Among the methylated sites, 76.84% were found within repeat sequences (Fig. 5d), significantly exceeding the 69.38% found in hAZFa repeat sequences. We further analyzed the distribution of repeat sequences by filtering the methylation level and found the enrichment of repeat sequences reinforced (Fig. 5e). Methylated genomic regions from scratch exhibited enrichment in intergenic regions, introns and promoters but showed less enrichment in exons (Fig. 5f).
a, DAPI (blue), 5mC (green) and 5hmC (red) are co-staining in mouse embryos (8 hours after injection). Scale bars, 20 µm. A magnified view shows the yeast nucleus in the cytoplasm. Scale bars, 1 µm. Quantification of 5mc and 5hmc staining (n = 10). b, The genome browser view shows DNA methylation of hAZFa in mouse one-cell embryos (three biological replicates) compared to AZFa in human sperm and zygote43. c, Numbers of CpG coverage sites and methylated sites of hAZFa in mouse one-cell embryos. d, The distribution of methylated sites in different transposable elements. e, The distribution of repeat sequences by filtering the methylation level of hAZFa in mouse one-cell early embryos (n = 3). Error bars, mean ± s.d. f, Bar charts showing the distribution of methylated sites in different genomic regions (n = 3). Error bars, mean ± s.d. g, Sequence feature of a 244-kb data-carrying BAC (up) and Integrative Genomics Viewer snapshot showing DNA methylation (two biological replicates) in mouse one-cell embryos (bottom). h, hours; LINE, long interspersed nuclear element; LTR, long terminal repeat; SINE, short interspersed nuclear element; UTR, untranslated region.
To confirm whether the cellular environment can identify any DNA sequence and introduce de novo DNA methylation, we injected a 244-kb data-carrying BAC44, which did not exist in nature. The sequence lacks basic features of the natural sequence (such as coding or regulatory information, homopolymer nucleotide runs and repeat frequency and position). It had 50% GC (G, guanine nucleotide; C, cytosine nucleotide) content and an average of 62 CpG sites per kilobase (Fig. 5g), which is 7.8 times than that of hAZFa. The random sequence DNA methylation level in one-cell embryos was only 0.9% (Extended Data Fig. 7d), with a 100% mapping coverage rate. Based on these data, we propose that de novo DNA methylation does not happen stochastically, is sequence dependent and is strongly enriched for repeat sequences.
Overall, we successfully delivered Mb-scale synthetic human DNA into mouse oocytes, and we observed the establishment of de novo mammalian epigenetic modifications, including nucleosome remodeling and DNA methylation, accompanied by 5hmC modification in mouse early embryos.
Gene expression in mouse early embryos
It was reported that many Y chromosome genes, including DDX3Y, are actively expressed in human eight-cell embryos when human zygotic genomic activation occurs45. Therefore, we next asked whether appropriate gene expression of hAZFa occurred in mouse early embryos and, if so, when it took place. We performed RNA-seq analysis at each of the one-cell, two-cell, four-cell and morula stages. We observed substantial expression of the coding genes DDX3Y and UTY and the human testis-specific long non-coding RNA (lncRNA) TTTY15 expressed at the mouse four-cell stage (Fig. 6a and Extended Data Fig. 8a). However, their expression levels were not equal, and the expression level showed considerable variation. The gene expression profile of hAZFa is similar to natural AZFa in human early embryos46, but the expression timing is more closely matched to that of homologous mouse genes in mouse early embryos. We further constructed and injected a 76-kb BAC containing full-length DDX3Y and TTTY15 into parthenogenic embryos. We observed the genes initiating transcription in early two-cell embryos with nearly equal expression levels (Extended Data Fig. 8b). Taken together, these findings indicate that the expression of hAZFa in mouse early embryos is regulated by the cellular environment.
a, Genome browser view showing RNA-seq signals of hAZFa genes in mouse early embryos compared to those of AZFa in human early embryos46 and homologous genes in wild-type mouse early embryos. b, Bar chart showing the DNA methylation of hAZFa (three biological replicates for one-cell and two-cell, and two biological replicates for four-cell). c, Genome browser view showing RNA-seq signals of whole hAZFa regions with or without 5-azadC (two-cell) and DMOG (four-cell and 12-hour delay) treatments. h, hours; TET, ten-eleven translocation.
Based on the limited presence of repressive histone marks, we speculate that DNA methylation plays a crucial role in regulating the expression of hAZFa at the four-cell stage. We observed that the number of methylated CpG sites in hAZFa significantly decreased from the one-cell stage to the four-cell stage, especially in the promoter region (Fig. 6b), indicating that demethylation occurred in hAZFa. Furthermore, we added 5-aza-deoxycytidine (5-azadC), an inhibitor of DNA methyltransferases (DNMTs), during parthenogenetic embryo culture and found that gene expression initiation occurred at the early two-cell stage (Fig. 6c). We added dimethyloxallyl glycine (DMOG), which efficiently blocks the activity of Tet enzymes in vitro47, during the parthenogenetic embryo culture and found that gene expression of hAZFa was effectively repressed (Fig. 6c). However, the scatter plot shows a slight developmental delay by treating DMOG (Extended Data Fig. 8c). We postponed the collection of four-cell embryos by 12 hours and verified that the suppression of gene expression on hAZFa was not due to developmental delay. Taken together, these data suggest that de novo DNA methylation plays a key role in regulating the expression of hAZFa at the four-cell stage.
Discussion
The epigenetic modifications on natural chromosomes are inherited and maintained48,49, making it difficult to erase the intrinsic epigenetic marks to study the basic principles of how it is established. Our SynNICE method lays the foundation by leveraging synthetic genomics to create a completely artificial genome that initially lacks DNA methylation and does not exist in nature. We observed that DNA methylation of synthetic, naive, Mb-scale human DNA from scratch did not occur randomly but was significantly enriched for repeat elements and intergenic regions. The observed pattern closely resembles the re-methylation changes (from less than 25% to more than 75%) observed in repeat elements during preimplantation development in human embryos43. The de novo DNA methylation of synthetic Mb-scale DNA is not promoted by H3K9me3, although DNA methylation and H3K9me3 frequently co-localize and act together to silence genes and repetitive elements. We speculated that the DNA-binding factors or lncRNA could recognize the DNA sequence and spontaneously methylate to repress their transcriptional activity and maintain stability. Our study first demonstrated the remodeling processes that regulated gene expression of synthetic Mb-scale human DNA at a temporal resolution. Unlike episomal plasmids, which are transcribed at the early two-cell stage, the synthetic Mb-scale DNA initially wrapped by yeast nucleosome is expressed in mouse four-cell embryos after the remodeling processes. We emphasize the important role of de novo DNA methylation in regulating the synthetic genome during early embryonic development. However, beyond epigenetic effects, the state of chromatin structure and regulation by other factors may also impact the expression of synthetic Mb-scale DNA.
In this work, we first accomplished the chemical synthesis and assembly of a Mb-scale continuous region of the human Y chromosome in yeast. The reference genome NCBI36/hg18 that we used for hAZFa demonstrates exceptionally high sequence identity (99.91%) with the current T2T CHM13v2.0/hs1 reference genome. We further assembled another human genome locus—the 498-kb human AZFc region (hg 38, chrY: 23,542,842–24,041,142)—from 100 short DNA fragments (Extended Data Fig. 9 and Supplementary Table 9). This result further validates our SynNICE method and demonstrates a methodological advancement for precise de novo assembly of large, highly repetitive human DNA sequences. Although hAZFa contains more repetitive sequences than the yeast genome, hAZFa in yeast remained stable for at least 50 generations in continuous passage culture. Even after 200 passages, seven of 10 random colonies stably maintained hAZFa (Extended Data Fig. 10a). Considering the euchromatin or heterochromatin environment in nuclei, inserting location highly affects the regulation results of our synthetic, naive genome. Thus, we prefer to use a host nuclei-independent system to study the function of de novo epigenetic modifications on hAZFa. By whole-genome sequencing (WGS) of mouse embryos, we have explicitly shown that the hAZFa in mouse early embryos can be stable until the morula stage (Extended Data Fig. 10b,c), which ensures the reliability of our system for studying the de novo remodeling and transcriptional regulation. However, we observed that the average depth of hAZFa was not halved during cell division (Extended Data Fig. 10d), indicating that hAZFa did not replicate in tandem with the mouse genome. Future research may reconstruct the hAZFa using the HAC vector in yeast50 and deliver it into mouse early embryos.
The SynNICE method is a major step forward in the challenge of delivering large, intact DNA molecules, especially into mammalian embryos. By using yeast nuclei, the method decouples the DNA size limitation from delivery efficiency. Without the use of crosslinking fixatives, the intact 1.14-Mb hAZFa retained a chromosomal structure in vitro, which can be stably stored for over 6 months at −80 °C. We speculate that our isolated nuclei could be as convenient to use as E. coli competent cells. We envision that such technology could be used to simultaneously deliver several different yeast nuclei containing different Mb-scale synthetic DNA fragments at a time, which will significantly accelerate the de novo construction of a synthetic mammalian chromosome (chromosome-level synthesis). The yeast nuclear membrane comprises two phospholipid bilayers, and the isolated nuclei were approximately 1 μm (ref. 51). By combining this advance with methods for membrane fusion and electroporation52,53, we anticipate that the yeast nuclei isolated by our technology could deliver Mb-scale human DNA to various cell types. Further optimization may involve efforts to inactivate the endogenous yeast chromosomes by CRISPR–Cas9 in vitro to prevent simultaneous entry and interference of endogenous yeast chromosomes into the cell54. Furthermore, facilitated by SynNICE, we could redesign additional variants of the human genome locus in yeast and test the genomic and epigenomic functions using our embryo model.
Our method demonstrates the precise assembly and delivery of synthetic Mb-scale DNA into mammalian embryos, leading to major implications for understanding the regulation and expression of synthetic genomes as well as for the development of future genome writing technologies. Further study on investigating the mechanisms of establishing epigenetic regulation from scratch will help to generate knowledge about epigenetic regulations and developmental biology in higher eukaryotes and move science toward more effective grand-scale engineering to address human health problems.
Methods
Strains and growth conditions
A full list of strains and plasmids used in this study can be found in Supplementary Tables 10 and 11. The plasmids were purified from E. coli cultures in Luria Broth supplemented with the appropriate antibiotic. Yeast cells were grown in either YPAD medium or Synthetic Complete (SC) medium lacking the indicated amino acids.
Synthesis and assembly of SynA, SynG, SynB and SynC
Twenty-three DNA fragments (40–71 kb) were assembled in S. cerevisiae strain BY4741 by standard LiAc/SS-DNA/PEG transformation protocol55. The DNA cassettes were pooled, digested with NotI and purified using a TIANgel Purification Kit (TIANGEN, DP209-03). Yeast colonies were selected on SC-His agar plates and incubated at 30 °C for 3 days. The assembled DNA fragments were digested by NotI and purified by ethanol precipitation.
SynA, SynG, SynB and SynC were assembled in S. cerevisiae VL6-48α or VL6-48a by the yeast protoplast transformation56. VL6-48α/VL6-48a cultures were grown in 50 ml of YPAD (optical density at 600 nm (OD600) = 0.2) at 30 °C for approximately 5 hours and then rinsed with a 1 M sorbitol. Cell pellets were resuspended in 20 ml of SPE solution containing 20 μl of zymolyase-20T (10 mg ml−1) and 40 μl of β-mercaptoethanol (Sigma-Aldrich, M3148) at 30 °C for 40 minutes. The protoplasts were then washed twice with 1 M sorbitol and resuspended in 200 μl of STC solution. For transformation, 50 ng of PCR-amplified linearized vector was mixed with the purified DNA fragments and added to the protoplast suspension, followed by 10-minute incubation. Then, 800 µl of PEG 8000 solution was added and incubated at room temperature for 10 minutes. Cells were resuspended in 800 µl of SOS solution and incubated at 30 °C for 40 minutes. The solution was transferred to a melted SORB-TOP selection medium and incubated at 30 °C for 5–7 days until colonies appeared.
Assembly of SynAG, SynBC and hAZFa
VL6-48α and VL6-48a strains harboring the preassembled fragments (SynA, SynB, SynG and SynC) were grown overnight in 3 ml of selective SC medium. Equal volumes (OD600 = 0.2) of each strain were mixed in 3 ml of YPAD and cultured for 8 hours. Serial 10-fold dilutions (100, 10−1 and 10−2) were plated on triple-auxotroph SC medium. After diploid formation, the SynA fragment was efficiently incorporated into the SynG–Ura3 fragment via CRISPR–Cas9-induced site-specific cleavage and yeast homologous recombination. For the final assembly of 1.14-Mb hAZFa, we improved the positive rate by screening for TRP1 marker loss through replica plating.
Yeast colony PCR and PFGE analysis
We used 24 primer pairs to verify junctions between each approximately 50-kb fragment in hAZFa and mating-type detection (Supplementary Table 12). Yeast chromosomal DNA was prepared using a CHEF Yeast Genomic DNA Plug Kit (Bio-Rad, 703593) and initially separated by 4 hours under the ‘5–430 kb’ program using the Pippin Pulse system (Sage Science). For restriction analysis, yeast agarose plug was digested with MluI (New England Biolabs (NEB), R3198L) for SynAG and SynBC, and with BsiWI (NEB, R3553) for hAZFa at 37 °C, in a 1% agarose gel (Bio-Rad, 161-3108), 0.5× TBE (Beyotime, ST718), 6 V cm−1, 60–120 seconds, running for 24 hours PFGE (CHEF MAPPER system; Bio-Rad) at 14 °C. For hAZFa analysis using AscI (NEB, R0558L) digestion, the yeast chromosome spreading step was excluded.
Total RNA extraction of Yeast_hAZFa
Total RNA was extracted from 25 ml of yeast cultured (OD600 = 1.0–1.2) using TRIzol reagent (Invitrogen, 15596018) following the manufacturer’s procedure. RNA integrity was verified by both Bioanalyzer 2100 (Agilent Technologies) analysis (RNA integrity number > 7.0) and denaturing agarose gel electrophoresis.
Yeast_hAZFa RNA-seq library preparation and sequencing
Poly(A) RNA was purified from 1 μg of total RNA using Dynabeads Oligo (dT)25 by using two rounds of purification. Then, the poly(A) RNA was fragmented into small pieces using a Magnesium RNA Fragmentation Module under 94 °C for 5–7 minutes. Next, the fragmented RNA was reverse transcribed to cDNA by SuperScript II Reverse Transcriptase (Invitrogen, 18064022), followed by synthesis of U-labeled second-stranded DNAs with E. coli DNA polymerase I, RNase H (NEB, M0297S) and dUTP Solution (Thermo Fisher Scientific, R0133). An A-base was then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Single-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads (Beckman Coulter, A63881). After the heat-labile UDG enzyme treatment of the U-labeled second-stranded DNAs, the ligated products were amplified with eight cycles of PCR. Qualified libraries were then loaded on an Illumina NovaSeq 6000 platform. HISAT2 version 2.1.0 was used to map clean reads to the yeast reference genome.
Yeast_hAZFa isoform sequencing library construction and sequencing
A SMARTer PCR cDNA Synthesis Kit (Takara, 634926) was used to reverse transcribe total RNA into cDNA. Isoform sequencing (Iso-seq) library preparation and sequencing were carried out by the ToFU method57. In brief, the Iso-seq library was constructed by using a SMRTbell Template Prep Kit (PacBio, 100-938-900) following the manufacturer’s procedure. The library quality and concentration were measured by Agilent 2100 Bioanalyzer and Qubit. Then, the PacBio binding kit was used to bind the library with primer and polymerase. Next, the final library was purified with AMpureXP beads, and Sequel II Sequencing Kit 2.0 Bundle (PacBio, 101-849-000) was used for sequencing. Raw reads were processed into error-corrected reads of insert (ROIs) using the Iso-seq pipeline with minFullPass = 3 and minPredictedAccuracy = 0.9. Then, full-length, non-chimeric transcripts were determined by searching for the poly(A) tail signal and the 5′ and 3′ cDNA primers in ROIs. Iterative clustering for error correction (ICE) was used to obtain consensus isoforms, and Quiver version 2.3.0 (ref. 58) was used to polish full-length consensus sequences from ICE. High-quality full-length transcripts were classified with the criteria of post-correction accuracy above 99%. Full-length consensus sequences were mapped to reference genome using GMAP version 2020-06-30 (ref. 59). Pbtranscript-ToFU package version 2.3.2 was used to collapse the mapped reads with min-coverage = 85% and min-identity = 90%.
Yeast_hAZFa proteome analysis
The strains were cultivated to OD600 = 1.0–1.2. After collection of the strains, yeast protein was extracted with 8 M urea mixed with 1% protease inhibitor (Sigma-Aldrich, P8340). Then, 200 μg of total protein was reduced by DTT at 30 °C for 60 minutes. Next, IAM was added. The free sulfhydryl group was blocked, and the light-avoidance reaction was performed for 45 minutes at room temperature. The proteins were precipitated with acetone and rinsed. TEAB was added, and the solution was sonicated on ice to facilitate protein dissolution. Trypsin was used to digest the proteins at 37 °C overnight. The peptides were desalted with a C18 SPE column (Phenomenex, 8B-S100-AAK), and the eluted peptides were drained with a vacuum concentrator. Then, 20 μg of peptides was pre-separated into 15 fractions by using Shimadzu LC20AD. Each fraction was analyzed for 1 hour using a mass spectrometry system (Thermo Fisher Scientific HF) with a loading volume of approximately 1 µg to establish a database. Next, mass spectrometry data acquisition was performed using the DIA method60. For proteomic analysis, Spectronaut version 13.4 and MaxQuant version 1.5.2.8 were used for mass spectrometry data analysis.
NICE
Yeast cells of the desired strains were grown to OD600 = 2. Cells were harvested by centrifugation at 600g and washed with water and 1 M sorbitol. Spheroplasts were prepared by resuspension in SPEM solution with β-mercaptoethanol and Zymolyase-100T (Nacalai Tesque, 07665-55). The spheroplast pellets were resuspended in sorbitol and washed with Ficoll (Cytiva, 17-0300-50) solution. Spheroplasts were lysed mechanically using a Polytron homogenizer in 8% PVP solution, and the lysis was monitored by microscopy with DAPI staining for DNA and WGA (Thermo Fisher Scientific, W11262) staining for the cell membrane. After homogenization, the lysate was purified by two-step sucrose (Sigma-Aldrich, V900116) density gradient centrifugation (2.5 M/1.875 M and 2.5 M/1.5 M) at 100,000g and 200,000g, respectively (Beckman Coulter, Optima XPN-100). The lysis and purification buffer was supplemented with 1× DNase inhibitor (Bestbio, BB-6106-1), 1× protease inhibitor (Sigma-Aldrich, P8340), 1× Solution P (0.2 mg ml−1 PMSF and 4 mg l−1 Pepstatin A), 10 mM EDTA-KOH (pH 8.0), 1 mM spermine (Sigma-Aldrich, S4264) and 2 mM spermidine (Sigma-Aldrich, 85558). The highly enriched nuclei were analyzed using PFGE, aliquoted into 20–30 tubes (10 μl each), flash frozen in liquid nitrogen and stored at −80 °C. For each experiment, a single aliquot was thawed on ice and used for microinjection within 4 hours.
ATAC–seq library preparation and sequencing
The strains of Yeast_hAZFa and Yeast_Control were cultivated to OD600 = 1.0–1.2. Samples were lysed in prechilled lysis buffer (15 mM Tris-HCl pH 7.5, 20 mM NaCl, 80 mM KCl, 0.5 mM spermine, 5 mM 2-ME and 0.2% Triton X-100) for 10 minutes at 4 °C. DNA fragmentation by Tn5 was performed according to a standard protocol61, and the final library was sequenced on the Illumina NovaSeq 6000. Adapters and low-quality reads were filtered out through Trimmomatic version 0.36. Clean reads were mapped to the reference genome by HISAT2. PCR duplicates were removed using SAMtools version 1.3.1, and MACS2 version 2.1.1 was used to call peaks. The final mapped BAM files were converted to the bigwig format using bamCoverage for visualization in Integrative Genomics Viewer version 2.16.2.
ChIP–seq library preparation and sequencing
Samples were crosslinked with 1% formaldehyde for 10 minutes at room temperature, and the reaction was quenched by the addition of glycine. A standard ChIP–seq protocol62 was carried out using an anti-H3K4me3 antibody (Cell Signaling Technology, 9751S) with 1:200 dilution and then sequencing on the Illumina NovaSeq 6000. Low-quality reads were removed using Trimmomatic. Clean reads were mapped to the reference genome by BWA version 0.7.15. SAMtools was used to remove potential PCR duplicates. MACS2 was used to call peaks. The mapped BAM files were finally converted to bigwig format via bamCoverage using reads per kilobase of transcript per million mapped reads (RPKM) normalization for visualization.
Hi-C library generation and sequencing
The method for preparing yeast nuclei libraries followed a similar procedure as that used for yeast cells but with additional optimization for crosslinking. In brief, yeast nuclei were fixed with 1% formaldehyde for 10 minutes at room temperature and quenched with 0.2 M glycine for 5 minutes. The yeast nuclei were then washed twice with PBS. Chromatin DNA was digested using 100 U of MboI (NEB), followed by the addition of biotin-14-dCTP (Invitrogen) and ligation using 50 U of T4 DNA ligase (NEB). After purification, DNA was sheared to 300–500-bp fragments. Ligation junctions were pulled down by using Dynabeads MyOne Streptavidin C1 (Thermo Fisher Scientific, 35002D). A Hi-C library was prepared with an NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7103L) according to the manufacturer’s instructions. Sequencing was performed on the Illumina NovaSeq 6000.
Construction of a contact map and 3D chromosome model from Hi-C data
The Hi-C data regarding Yeast_hAZFa and isolated nuclei were mapped iteratively to a combination of S. cerevisiae S288C genome and hAZFa sequence using ICE software package version 1f8815d0cc9e. After filtering out dangling ends and other unusable data, the valid pairs were binned into 10-kb non-overlapping genomic intervals to generate contact maps. The contact maps were normalized using an iterative method to eliminate systematic biases. The interactions across the whole genome of isolated nuclei compared to Yeast_hAZFa were transformed into z-scores using E. Crane’s method. z-scores were calculated for each 10 kb-bin to generate a standardized symmetric matrix by using the cworld toolkit (https://github.com/dekkerlab/cworld-dekker) ‘matrix2loess.pl’ script using the following parameters: --ca 0.005 --ez--ca and --ez ignore 0 s in all calculations63. z-scores of isolated nuclei were subtracted from those of Yeast_hAZFa to estimate the difference in their whole-genome interactions. 3D chromosomal structures were inferred using the Pastis version 0.1 method with the MDS model. The 3D model was constructed using the 10-kb contact maps. The rDNA region was excluded due to its high repeat content, which introduced significant noise in intrachromosomal interactions64.
Mouse oocyte collection, parthenogenetic activation, embryo culture and yeast nuclei injection
All animal maintenance and experimental procedures used in this study were carried out according to the Institutional Animal Care and Use Committee guidelines of Tianjin University (ethical approval protocol no. TJUE-2023-159). Four-week-old female C57BL/6 mice (purchased from Vital River) were injected intraperitoneally with PMSG (10 IU) and HCG (10 IU) 48 hours apart. MII oocytes were collected in M2 medium (Sigma-Aldrich, M7167) 13 hours after HCG. For yeast nuclei injection, a 10-μl aliquot of yeast nuclei was retrieved, kept on ice and injected using a Piezo impact-driven micromanipulator. MII oocytes were activated by treatment with 10 mM SrCl2 in the CZB medium containing 5 μg ml−1 cytochalasin B (Sigma-Aldrich, C2743). The injected oocytes were incubated in KSOM medium (Millipore, MR-121-D) for 30 minutes before activation. Five hours after activation, the embryos were then cultured in KSOM medium. The parthenogenetic one-cell, two-cell, four-cell and morula embryos were collected at 8, 26, 50 and 74 hours after injection, respectively.
Inhibition of Tet proteins was performed by adding 1 mM DMOG (Sigma-Aldrich, D3695) to the activation medium for 5 hours and to the KSOM medium for 3 hours, followed by culture in the KSOM medium until the four-cell stage. Inhibition of DNMT proteins was performed by adding 1 μM 5-azadC (Sigma-Aldrich, A3656) to the activation medium for 5 hours and to the KSOM medium until the two-cell stage.
For early embryo collection, C57BL/6 female mice were mated with PWK/PhJ males (purchased from The Jackson Laboratory) after HCG administration. One-cell, two-cell, four-cell and morula embryos were collected at 26, 35, 56 and 80 hours after HCG, respectively.
5mC and 5hmC straining of parthenogenetic embryos
Fixed embryos were treated with 4 N HCl for 10 minutes at room temperature, followed by neutralization in 100 mM Tris-HCl (pH 8.0) for 10 minutes. After 1-hour blocking in 2% BSA-PBS, embryos were incubated overnight at 4 °C with anti-5mC (Diagenode, C15200081) 1:100 and anti-5hmc (Active Motif, 39769) 1:1,000 primary antibody. After two rinses with PBS, samples were incubated with secondary antibodies Alexa Fluor 488–conjugated anti-mouse and Alexa Fluor 568–conjugated anti-rabbit (Invitrogen, A-21202 and A-11036, 1:1,000 diluted) for 1 hour at room temperature. After two rinses with PBS, samples were directly deposited on glass slides, mounted with VECTASHIELD medium containing DAPI (Vector Laboratories, H-1200) and covered by coverslips (Zeiss, 474030-9000-000). The images were taken with an LSM 900 confocal microscope system (Zeiss). Immunofluorescence images were analyzed with ImageJ version 1.53k.
Embryos RNA-seq library preparation and sequencing
All RNA-seq libraries were generated following the Smart-seq2 protocol65. The zona pellucida was gently removed by treatment with Tyrode’s solution (Sigma-Aldrich, T1788). Embryos were washed in M2 medium and then lysed in 2 μl of lysis buffer containing RNase inhibitor. Then, we carried out the amplification by the Smart-seq2 method. Qualified libraries were then loaded on an Illumina HiSeq platform for PE150 sequencing. Paired-end RNA-seq reads were trimmed and then mapped to the reference genome by TopHat version 2.0.12. HTSeq version 0.6.0 was used to calculate the counts of each gene, and fragments per kilobase of transcript per million mapped reads (FPKM) was used to assess the expression level. Integrative Genomics Viewer was used to view the mapping results.
WGBS library preparation and sequencing
Embryos were collected and lysed by incubation at 37 °C for 1 hour. Bisulfite conversion was performed on cell lysates using an EZ DNA Methylation-Gold Kit (Zymo Research, D5006). The purified libraries were prepared for 125-bp paired-end sequencing on an Illumina HiSeq 2500. All bisulfite sequencing reads were first trimmed to remove adaptors and low-quality bases using Trimmomatic. All the reads that passed quality control were mapped to the reference genome using Bismark version 0.16.3. Only reads with a unique mapping location in the genome were retained for further analysis. The DNA methylation level was determined by the ratio of the number of reads supporting C (methylated) to that of total reads (methylated and unmethylated), which was produced by the Bismark toolkit of bismark_methylation_extractor. The read coverage threshold used to call the DNA methylation level for any cytosine was 1× for each sample.
CUT&Tag library preparation and sequencing
CUT&Tag was performed following the protocol with minor modifications66. The zona pellucida was removed with Tyrode’s solution, and the polar body was removed with a sharp glass pipette. Samples were then resuspended by 100 μl of washing buffer. Then, 10 μl of Concanavalin-coated magnetic beads (Polysciences, 86057) for each sample were gently washed, resuspended by binding buffer and carefully added to the samples. The samples were then incubated at 23 °C for 10 minutes on a thermomixer (Eppendorf) at 400 r.p.m. After magnetic separation, samples were incubated overnight at 4 °C with H3K9me3 antibody (Active Motif, 39161) diluted at a ratio of 1:50 in antibody buffer, followed by secondary antibody (Invitrogen, SA5-10228) to a final concentration of 1:100 at 4 °C on a thermomixer for 3 hours at 400 r.p.m. The samples were washed twice in DIG washing buffer, and then 0.5 μl of pA-Tn5 was added with 100 μl of DIG-300 buffer. Samples were incubated at room temperature at 4 °C on the thermomixer for 1 hour at 400 r.p.m. and washed with 200 μl of DIG-300 buffer twice at the magnetic stand. Add 62.5 µl of tagmentation buffer (DIG-300 buffer plus 10 mM MgCl2) into every tube. Incubate at 37 °C for 1 hour. Add 1 µl of 20% SDS and incubate at 55 °C for 10 minutes. Add 200 µl of SPRI beads at room temperature for 15 minutes. Place the tubes on a magnet stand to clear and withdraw the liquid, add 800 µl of 80% ethanol to wash and repeat once. Place on the magnet and remove the liquid. Allow the tube to dry for 3 minutes and resuspend pellets in 29 µl of ddH2O. PCR was performed to amplify the libraries. All libraries were sequenced by Illumina HiSeq 2500 according to the manufacturer’s instructions. Trimmomatic was used to filter out low-quality reads. Clean reads were mapped to our reference genome by BWA. SAMtools was used to remove potential PCR duplicates and set stringent filtering on the alignment quality score (MAPQ = 42). MACS2 software was used to call peaks by default parameters.
WGS library preparation and sequencing
The number of one-cell, two-cell, four-cell and morula embryos collected for WGS was 20, 20, 10 and 8, separately. After cell lysis, multiple displacement amplification (MDA) was performed using a REPLI-g Advanced DNA Single Cell Kit (Qiagen). DNA libraries were performed by MGISEQ-2000RS High-throughput Sequencing Set (PE150) according to the manufacturer’s instructions. Sequencing was performed on the MGISEQ-2000 (MGI). Low-quality reads were trimmed out using SOAPnuke version 2.2.6 using the following parameters: -n 0.001 -l 10 -q 0.5 -Q 2. Clean reads were mapped to the reference genome by BWA version 0.7.17, and SAMtools was used to change the alignment result to BAM format. Qualimap version 2.2.1 was used to calculate coverage, sequencing depth and plotting of the alignment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available within the article and its supplementary tables. All data have been deposited at the Sequence Read Archive with BioProject number PRJNA1045349 and at the Gene Expression Omnibus with accession number GSE250094. Published data used in this study are as follows: RNA-seq of human early embryos, GSE36552; DNA methylation of human zygote and sperm, GSE81233; H3K9me3 ChIP–seq of mouse embryos, CRA004123; and H3K9me3 CUT&RUN of human embryos, HRA001391. Source data are provided with this paper.
References
Cello, J., Paul, A. V. & Wimmer, E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297, 1016–1018 (2002).
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010).
Fredens, J. et al. Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518 (2019).
Richardson, S. M. et al. Design of a synthetic yeast genome. Science 355, 1040–1044 (2017).
Wu, Y. et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science 355, eaaf4706 (2017).
Xie, Z.-X. et al. ‘Perfect’ designer chromosome V and behavior of a ring derivative. Science 355, eaaf4704 (2017).
Chari, R. & Church, G. M. Beyond editing to writing large genomes. Nat. Rev. Genet. 18, 749–760 (2017).
Pinglay, S. et al. Synthetic regulatory reconstitution reveals principles of mammalian Hox cluster regulation. Science 377, eabk2820 (2022).
Zhang, W. et al. Mouse genome rewriting and tailoring of three important disease loci. Nature 517, 321–326 (2015).
Brosh, R. et al. A versatile platform for locus-scale genome rewriting and verification. Proc. Natl Acad. Sci. USA 118, e2023952118 (2021).
Boeke, J. D. et al. The Genome Project-Write. Science 353, 126–127 (2016).
Lienert, F. et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 43, 1091–1097 (2011).
Borgel, J. et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100 (2010).
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016).
Lartigue, C. et al. Genome transplantation in bacteria: changing one species to another. Science 317, 632–638 (2007).
Zürcher, J. F. et al. Continuous synthesis of E. coli genome sections and Mb-scale human DNA assembly. Nature 619, 555–562 (2023).
Li, L. & Blankenstein, T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast–embryonic stem cell fusion. Nat. Protoc. 8, 1567–1582 (2013).
Fang, G. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239 (2012).
Montoliu, L. Large-scale preparation of yeast agarose plugs to isolate yeast artificial chromosome DNA. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot093955 (2018).
James, J. S., Dai, J., Chew, W. L. & Cai, Y. The design and engineering of synthetic genomes. Nat. Rev. Genet. 26, 298–319 (2025).
Zhang, W. et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science 355, eaaf3981 (2017).
Mitchell, L. A. et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science 355, eaaf4831 (2017).
Shen, Y. et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 355, eaaf4791 (2017).
Mitchell, L. A. et al. De novo assembly and delivery to mouse cells of a 101 kb functional human gene. Genetics 218, iyab038 (2021).
Jakobovits, A. et al. Germ-line transmission and expression of a human-derived yeast artificial chromosome. Nature 362, 255–258 (1993).
Brown, D. M. et al. Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic Acids Res. 45, e50 (2017).
Kachroo, A. H. et al. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921–925 (2015).
Hughes, J. F. & Page, D. C. The biology and evolution of mammalian Y chromosomes. Annu. Rev. Genet. 49, 507–527 (2015).
Skaletsky, H. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Sun, C. et al. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum. Mol. Genet. 9, 2291–2296 (2000).
Kennedy, B. K. Mammalian transcription factors in yeast: strangers in a familiar land. Nat. Rev. Mol. Cell Biol. 3, 41–49 (2002).
Gvozdenov, Z., Barcutean, Z. & Struhl, K. Functional analysis of a random-sequence chromosome reveals a high level and the molecular nature of transcriptional noise in yeast cells. Mol. Cell 83, 1786–1797.e5 (2023).
Niepel, M., Farr, J. C., Rout, M. P. & Strambio-De-Castillia, C. Rapid isolation of functionally intact nuclei from the yeast Saccharomyces. Preprint at bioRxiv https://doi.org/10.1101/162388 (2017).
Schreiner, S. M., Koo, P. K., Zhao, Y., Mochrie, S. G. & King, M. C. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun. 6, 7159 (2015).
Woo, E.-J. et al. Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol. Cell 14, 531–539 (2004).
Pasero, P., Braguglia, D. & Gasser, S. M. ORC-dependent and origin-specific initiation of DNA replication at defined foci in isolated yeast nuclei. Genes Dev. 11, 1504–1518 (1997).
Dang, Y., Yadav, R. P. & Chen, X. in Chromatin Accessibility: Methods and Protocols (eds Marinov, G. K. & Greenleaf, W. J.) 285–291 (Springer, 2023).
Kornberg, R. D. & Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999).
Armache, A. et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 583, 852–857 (2020).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
Jamieson, K., Rountree, M. R., Lewis, Z. A., Stajich, J. E. & Selker, E. U. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc. Natl Acad. Sci. USA 110, 6027–6032 (2013).
Buitrago, D. et al. Impact of DNA methylation on 3D genome structure. Nat. Commun. 12, 3243 (2021).
Zhu, P. et al. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 50, 12–19 (2018).
Chen, W. et al. An artificial chromosome for data storage. Natl Sci. Rev. 8, nwab028 (2021).
Ditton, H., Zimmer, J., Kamp, C., Rajpert-De Meyts, E. & Vogt, P. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 13, 2333–2341 (2004).
Yan, L. et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013).
Amouroux, R. et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233 (2016).
Margueron, R. & Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296 (2010).
Feng, S., Jacobsen, S. E. & Reik, W. Epigenetic reprogramming in plant and animal development. Science 330, 622–627 (2010).
Gambogi, C. W. et al. Efficient formation of single-copy human artificial chromosomes. Science 383, 1344–1349 (2024).
Taddei, A. & Gasser, S. M. Structure and function in the budding yeast nucleus. Genetics 192, 107–129 (2012).
Wickner, W. & Schekman, R. Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 (2008).
Tsong, T. Y. Electroporation of cell membranes. Biophys. J. 60, 297–306 (1991).
Shao, Y. et al. Creating a functional single-chromosome yeast. Nature 560, 331–335 (2018).
Gietz, R. D. & Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Kouprina, N. & Larionov, V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 3, 371–377 (2008).
Gordon, S. P. et al. Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS ONE 10, e0132628 (2015).
Chin, C.-S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Wu, T. D. & Watanabe, C. K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Lin, L. et al. High throughput and accurate serum proteome profiling by integrated sample preparation technology and single-run data independent mass spectrometry analysis. J. Proteom. 174, 9–16 (2018).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Kim, T. H. & Dekker, J. ChIP–quantitative polymerase chain reaction (ChIP–qPCR). Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot082628 (2018).
Maass, P. G., Barutcu, A. R., Weiner, C. L. & Rinn, J. L. Inter-chromosomal contact properties in live-cell imaging and in Hi-C. Mol. Cell 69, 1039–1045 (2018).
Lajoie, B. R., Dekker, J. & Kaplan, N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods 72, 65–75 (2015).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Acknowledgements
We thank Y. Li from the University of California, San Diego, Y. Zhao from Westlake University and S. Ye from Tianjin University for discussion and comments during the hAZFa study and preparation of the manuscript. We thank F. Zhong and H. Liu from the Yuan Laboratory for collecting mouse oocytes. We thank C. Liu from IOZ for advice on the CZB medium and embryo culture. We thank F. Gao for advice on data analysis and sequence design. We thank W. Dong from Zeiss China for advice on live images. This work was funded by the National Key R&D Program of China (2017YFC1200704) (Y.Y.) and the National Natural Science Foundation of China (21621004) (Y.Y.).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.L., Z.L. and Y.Y. Methodology: Y.L. developed and optimized the SynNICE method; J.Z. performed the assembly and PFGE; Y.L. and Z.L. performed the injection to mouse MII oocytes; X.H. prepared the CUT&Tag libraries; and D.L., L.Y., X.S. and X.J. advised on the design of hAZFa. Visualization: Y.L. performed all the images under the instruction of Z.L., and J.Z. performed yeast nuclei ATAC–see. Funding acquisition: Y.Y. Project administration: Y.Y. Supervision: Y.Y., Z.L., W.X. and L.Y. Writing—original draft: Y.L. Writing—review and editing: Y.Y., Y.L. and J.Z., with help from all authors.
Corresponding author
Ethics declarations
Competing interests
Y.Y., Y.L., J.Z. and Z.L. are co-investors on three patents related to SynNICE. L.Y. is co-founder and CEO of Qihan Biotech. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Giovanni Stracquadanio and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Lei Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Whole-genome sequencing of two AZFa microdeletion patients.
The genome browser view shows that both patients had ~798 kb deletions in the AZFa region.
Extended Data Fig. 2 Analysis of repeat sequence features of hAZFa compared with S. cerevisiae and E. coli genomes.
a, The percentage of each repeat class on hAZFa, S. cerevisiae, and E. coli. b, Total repetitive sequence content in hAZFa, S. cerevisiae and E. coli. c, Quantitation of repetitive element classes in hAZFa.
Extended Data Fig. 3 Assembly and characterization of hAZFa.
a, Restriction digestion analysis of SynA, SynB, SynG, and SynC followed by PFGE. Left, NruI enzyme digestion of SynG and λ PFG ladder as a marker. Right, SalI enzyme digestion of SynB, MluI enzyme digestion of SynA, SynC, and λ PFG ladder as a marker. n = 3 biological replicates. b, Restriction digestion analysis of SynAG and SynBC followed by PFGE. Left, MluI enzyme digestion of 599 kb SynAG and λ PFG ladder as a marker. Right, MluI enzyme digestion of 544 kb SynBC and λ PFG ladder as a marker. n = 3 biological replicates. c, Left, the sequencing coverage track for the full length of hAZFa was determined by using next-generation sequencing (NGS). The average depth of coverage was approximately 400×. Right, compared with the designed hAZFa locus, Yeast_hAZFa had 41 SNPs, 20 Indels, one intra-chromosomal translocation (ITX) of 115 bp, and no large structural variants (SVs).
Extended Data Fig. 4 Multi-omic characterization of hAZFa in S. cerevisiae.
a, Genome browser view showing expression of hAZFa have transcripts in yeast. b, Proteomic enrichment analysis of Yeast_hAZFa compared with Yeast_Control, with circle sizes representing protein number and color intensity indicating significance level. c, Total RNA yields from Yeast_hAZFa were similar to those from Yeast_Control. n = 3 biological replicates. d, Whole-protein extracts of Yeast_Control and Yeast_hAZFa were run on a 12% SDS-Bis-Tris acrylamide gel and stained with Coomassie blue, using NEB #P7712 as the protein marker. n = 3 biological replicates. e, 3D structure model shows interactions of hAZFa and endogenous chromosomes in yeast. f, Z-score difference contact heatmap shows a comparison of chromosome interactions of Yeast_hAZFa and Yeast_Control. Bin length, 10 kb; red and blue show increased and decreased chromatin interactions, respectively. g, Comparison of the endogenous yeast centromere relative interaction frequency between Yeast_hAZFa and Yeast_control. Data are presented as the mean ± SE. Two-tailed t-tests were used to evaluate differences between groups. (***) P < 0.001.
Extended Data Fig. 5 hAZFa in isolated yeast nuclei.
a, BsiWI enzyme digestion analysis of hAZFa in isolated nuclei followed by PFGE. n = 3 biological replicates. b, Southern blot to detect the integrity of hAZFa in isolated nuclei. Left, the original PFGE images of Fig. 3c. Middle, two independent southern blot experiments with four times NICE. Right, the ratio of full-sized hAZFa in isolated nuclei by NICE, compared with in Yeast_hAZFa calculated from southern blot out of PFGE (n = 4). c, Pulsed-field gel electrophoresis confirmed that the chromosomal DNA within the nuclei remained intact after 193 days of frozen storage at –80 °C. n = 3 biological replicates. d, Average FPKM of ATAC-seq peaks of each chromosome (up) and raw peaks on hAZFa (bottom), including hAZFa in Yeast_Control, Yeast_hAZFa, and isolated Yeast nuclei_hAZFa. e, Representative fluorescence images of ATAC-see (green) in isolated nuclei. Right, Violin plot of signal intensity of isolated nuclei. Sample sizes are as follows: Yeast nuclei_ hAZFa, n = 100. f, Average FPKM of H3K4me3 ChIP-seq peaks for each chromosome, including hAZFa in Yeast_Control, Yeast_hAZFa, and isolated Yeast nuclei_hAZFa. g, Representative fluorescence images depicting H3K4me3 in yeast and isolated nuclei. Right, violin plot of signal intensity of yeast and isolated nuclei. Sample sizes are as follows: Yeast_hAZFa, n = 100 and Yeast nuclei_hAZFa, n = 100.
Extended Data Fig. 6 3D structures of Yeast nuclei_hAZFa compared with Yeast_ hAZFa.
a, 3D formation shows interactions of hAZFa and endogenous chromosomes in isolated nuclei. b, Z-score difference contact heatmap shows a comparison of chromosome interactions of Yeast nuclei_hAZFa and Yeast_hAZFa. Bin length, 10-kb; red and blue show increased and decreased chromatin interactions, respectively. c, Comparison of the centromere relative interaction frequency between isolated yeast nuclei and Yeast_hAZFa. Data are presented as the mean ± s.e. Two-tailed t-tests were used to evaluate differences between groups. (***) P < 0.001.
Extended Data Fig. 7 H3K9me3 signals of hAZFa, and DNA methylation of hAZFa and data-carrying BAC in mouse early embryos.
a, Genome browser view showing that de novo H3K9me3 of hAZFa in mouse 2-cell embryos compared with AZFa in human 4-cell and 8-cell embryos. b, The random snapshot from UCSC Genome Browser showing the H3K9me3 profile of our injected mouse early two-cell embryos performed by CUT&Tag, in comparison to the published H3K9me3 Chip-seq data from wild-type mouse early 2-cell embryos. c, The DNA methylation levels of hAZFa and yeast genome in mouse one-cell embryos compared with those in yeast. d, The DNA methylation level of data-carrying BAC in E. coli and in mouse one-cell embryos.
Extended Data Fig. 8 Transcriptional regulation of 76-kb BAC and hAZFa.
a, Genome browser view showing RNA-seq signals of the DDX3Y genes on hAZFa in mouse four-cell and morula embryos. b, Left, construction and microinjection of 76-kb BAC containing full-length TTTY15 and DDX3Y genes with flanking 5-kb regions. Right, RNA-seq of TTTY15 and DDX3Y genes on BAC in one-cell and early two-cell mouse embryos. c, Scatter plot showing gene expression fold-change upon adding 5-azadC, DMOG, and DMOG with 12 h postponing (2 biological replicates). FC, fold-change. Red lines, local regression fitting.
Extended Data Fig. 9 De novo assembly of the 498-kb human AZFc region using SynNICE.
a, 498 kb human AZFc region schematic. b, Restriction digestion analysis of ten 50-kb DNA fragments followed by PFGE. NotI digestion and λ DNA-Mono Cut Mix (NEB, N3019S) as a marker. n = 3 biological replicates. c, Restriction digestion analysis of 249.4 kb DNA fragments C51-100 and C101-150 followed by PFGE. MluI digestion and λ PFG ladder (NEB, N0341S) as a marker. n = 2 biological replicates. d, Restriction digestion analysis of 498 kb DNA fragment C51-150 followed by PFGE. MluI digestion (left) and BsiWI digestion (right), λ PFG ladder as a marker. n = 2 biological replicates.
Extended Data Fig. 10 Stability analysis of hAZFa in yeast and in mouse early embryos.
a, The ratio of strains containing the correct hAZFa constructs for 50, 100, 150, and 200 passages verified by PFGE. n = 3 biological replicates. Error bars, mean ± SD. b and c, The WGS coverage and IGV snapshot of hAZFa in mouse one-cell, two-cell, four-cell, and morula embryos. n = 2 biological replicates. d, The average depth of hAZFa in mouse one-cell, two-cell, four-cell, and morula embryos (average depth of each sample: ranging from 39.367 to 39.753). n = 2 biological replicates.
Supplementary information
Supplementary Information
Supplementary protocols of ‘Protein analysis and SDS-PAGE’, ‘Yeast Immunofluorescence’, ‘ATAC–see for isolated yeast nuclei’ and ‘Southern blot analysis’ removed from the Methods section. A guide to Supplementary Tables 1–12 (tables supplied separately).
Supplementary Table
Descriptions are provided in the Supplementary Information file.
Source data
Source Data
Unprocessed gels.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Zhou, J., Liu, D. et al. De novo assembly and delivery of synthetic megabase-scale human DNA into mouse early embryos. Nat Methods 22, 1686–1697 (2025). https://doi.org/10.1038/s41592-025-02746-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02746-8
This article is cited by
-
Synthetic rewriting technologies in mammalian cells
Nature Communications (2026)