Abstract
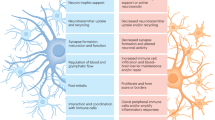
Traditional approaches to studying astrocyte heterogeneity have mostly focused on analyzing static properties, failing to identify whether subtypes represent intermediate or final states of reactive astrocytes. Here we show that previously proposed neuroprotective and neurotoxic astrocytes are transitional states rather than distinct subtypes, as revealed through time-series multiomic sequencing. Neuroprotective astrocytes are an intermediate state of the transition from a nonreactive to a neurotoxic state in response to neuroinflammation, a process regulated by the mTOR signaling pathway. In Alzheimer’s disease (AD) and aging, we observed an imbalance in neurotoxic and neuroprotective astrocytes in animal models and human patients. Moreover, targeting mTOR in astrocytes with rapamycin or shRNA mitigated astrocyte neurotoxic effects in neurodegenerative mouse models. Overall, our study uncovers a mechanism through which astrocytes exhibit neuroprotective functions before becoming neurotoxic under neuroinflammatory conditions and highlights mTOR modulation specifically in astrocytes as a potential therapeutic strategy for neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The bulk RNA-seq and scRNA-seq raw sequencing data for MD-astrocytes, as well as the bulk RNA-seq raw sequencing data for SFM-astrocytes generated in this study, have been deposited and are publicly available at the Genome Sequence Archive under accession numbers CRA011193 and CRA020575, respectively, in the National Genomics Data Center database under project ID PRJCA017165. The mass spectrometry proteomics data have been deposited to the OMIX under accession number OMIX004203 and project ID PRJCA017165. These proteomics data have also been submitted to the ProteomeXchange Consortium via the PRIDE48 partner repository with the dataset identifiers PXD058122, PXD058125, PXD058126 and PXD058128, which will be made publicly available upon publication. The publicly available bulk RNA-seq and snRNA-seq datasets used in this study are available at the Genome Sequence Archive under accession number CRA008312, at the Gene Expression Omnibus database under accession numbers GSE137028, GSE148612, GSE148610, GSE140511, GSE185553, GSE185277 and GSE198323, as well as at The Rush Alzheimer’s Disease Center Research Resource Sharing Hub at https://www.radc.rush.edu/docs/omics.htm (snRNA-seq PFC) or at Synapse (https://www.synapse.org/#!Synapse:syn18485175) at https://doi.org/10.7303/syn18485175.
Code availability
This paper does not report original code. Published or publicly available software, tools, algorithms and packages are cited with their version numbers or links in Methods and Reporting Summary.
References
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Guttenplan, K. A. et al. Knockout of reactive astrocyte activating factors slows disease progression in an ALS mouse model. Nat. Commun. 11, 3753 (2020).
Guttenplan, K. A. et al. Neurotoxic reactive astrocytes drive neuronal death after retinal injury. Cell Rep. 31, 107776 (2020).
Yun, S. P. et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 24, 931–938 (2018).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Lee, E. et al. A distinct astrocyte subtype in the aging mouse brain characterized by impaired protein homeostasis. Nat. Aging 2, 726–741 (2022).
Chen, Z. P. et al. Lipid-accumulated reactive astrocytes promote disease progression in epilepsy. Nat. Neurosci. 26, 542–554 (2023).
Michelucci, A., Heurtaux, T., Grandbarbe, L., Morga, E. & Heuschling, P. Characterization of the microglial phenotype under specific pro-inflammatory and anti-inflammatory conditions: effects of oligomeric and fibrillar amyloid-beta. J. Neuroimmunol. 210, 3–12 (2009).
Mills, C. D., Kincaid, K., Alt, J. M., Heilman, M. J. & Hill, A. M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164, 6166–6173 (2000).
Sala Frigerio, C. et al. The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27, 1293–1306 (2019).
Masuda, T. et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019).
Jakel, S. et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547 (2019).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271 (2019).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Marques, S. et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016).
Sadick, J. S. et al. Astrocytes and oligodendrocytes undergo subtype-specific transcriptional changes in Alzheimer’s disease. Neuron 110, 1788–1805 (2022).
McCarthy, K. D. & de Vellis, J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 85, 890–902 (1980).
Guttenplan, K. A. et al. Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature 599, 102–107 (2021).
Zhang, L. et al. Alleviating symptoms of neurodegenerative disorders by astrocyte-specific overexpression of TMEM164 in mice. Nat. Metab. 5, 1787–1802 (2023).
Foo, L. C. et al. Development of a method for the purification and culture of rodent astrocytes. Neuron 71, 799–811 (2011).
Hasel, P., Rose, I. V. L., Sadick, J. S., Kim, R. D. & Liddelow, S. A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 24, 1475–1487 (2021).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Clarke, L. E. et al. Normal aging induces A1-like astrocyte reactivity. PNAS 115, E1896–E1905 (2018).
Zhou, Y. et al. Molecular landscapes of human hippocampal immature neurons across lifespan. Nature 607, 527 (2022).
Pan, J., Ma, N., Yu, B., Zhang, W. & Wan, J. Transcriptomic profiling of microglia and astrocytes throughout aging. J. Neuroinflamm. 17, 97 (2020).
Zhang, Z. et al. Brain-restricted mTOR inhibition with binary pharmacology. Nature 609, 822–828 (2022).
He, D. et al. Activation of HCA2 regulates microglial responses to alleviate neurodegeneration in LPS-induced in vivo and in vitro models. J. Neuroinflamm. 20, 86 (2023).
Han, X. et al. Small molecule-driven NLRP3 inflammation inhibition via interplay between ubiquitination and autophagy: implications for Parkinson disease. Autophagy 15, 1860–1881 (2019).
Mendes-Oliveira, J., Lopes Campos, F., Videira, R. A. & Baltazar, G. GPER activation is effective in protecting against inflammation-induced nigral dopaminergic loss and motor function impairment. Brain Behav. Immun. 64, 296–307 (2017).
Hoban, D. B. et al. Further characterisation of the LPS model of Parkinson’s disease: a comparison of intra-nigral and intra-striatal lipopolysaccharide administration on motor function, microgliosis and nigrostriatal neurodegeneration in the rat. Brain Behav. Immun. 27, 91–100 (2013).
Herrera, A. J., Castano, A., Venero, J. L., Cano, J. & Machado, A. The single intranigral injection of LPS as a new model for studying the selective effects of inflammatory reactions on dopaminergic system. Neurobiol. Dis. 7, 429–447 (2000).
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325 (2021).
Cunningham, C., Dunne, A. & Lopez-Rodriguez, A. B. Astrocytes: heterogeneous and dynamic phenotypes in neurodegeneration and innate immunity. Neuroscientist 25, 455–474 (2019).
Hasel, P. et al. Molecular and metabolic heterogeneity of astrocytes and microglia. Cell Metab. 35, 555–570 (2023).
Qiu, J. et al. Single-cell RNA-seq reveals heterogeneity in metastatic renal cell carcinoma and effect of anti-angiogenesis therapy in the pancreas metastatic lesion. Cancer Lett. 601, 217193 (2024).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Meier, F. et al. diaPASEF: parallel accumulation-serial fragmentation combined with data-independent acquisition. Nat. Methods 17, 1229–1236 (2020).
Zhang, L., Li, J. & Lin, A. Assessment of neurodegeneration and neuronal loss in aged 5XFAD mice. STAR Protoc. 2, 100915 (2021).
Beaudoin, G. M. et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 7, 1741–1754 (2012).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Barbar, L. et al. CD49f is a novel marker of functional and reactive human iPSC-derived astrocytes. Neuron 107, 436–453 (2020).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Acknowledgements
We thank M.-m. Poo for helpful discussions and insightful comments. We are grateful to members of the Optical Imaging facility (W. Qian, Y. Zhang and Q. Hu), as well as members of the FACS facility (H. Wu and L. Quan), for technical assistance. This study was supported by the STI2030-Major Projects of China (2021ZD0200900, H.Z.), the National Key Research and Development Program of China (2022YFC3400100, H.Z.), the Shanghai 2023 Special Biopharmaceutical Science and Technology Support Projects (23S41900300, H.Z.), the National Natural Science Foundation of China (82001202, B.W.) and the Shanghai Pujiang Talent Program (2020PJD006, B.W.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
L.Z. and H.Z. conceived this work. L.Z., Z.X., Z.J. and S.C. carried out the study. L.Z. performed primary astrocyte culture, immunofluorescence staining, immunoblotting and qPCR assays, prepared samples for proteomics/RNA-seq/scRNA-seq, analyzed data, and wrote the paper, with valuable revision from all of the authors. Z.X. performed the bioinformatic data analysis. Z.J. performed primary astrocyte culture, flow cytometry assay and qPCR assay, analyzed the data, and wrote the paper. S.C. constructed vectors, performed qPCR assays and analyzed the data. X.H. prepared AAVs. Q.W. and X.L. conducted AAV and drug injections on the LPS-induced PD and 5XFAD models. T.B. contributed to neuronal toxicity experiments. Y.C. contributed to immunofluorescence staining and qPCR assay. T.L., B.W. and J.Z. contributed to immunofluorescence staining, immunoblotting assays and culture of SFM-astrocytes. Z.L. contributed to results discussions and paper preparation. H.Z. supervised the project and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
H.Z. is a founder and scientific adviser of Genemagic Biosciences. The other authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Won-Suk Chung, Blanca Díaz Castro and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Evaluation of the stability of neurotoxic and neuroprotective substates in response to inflammatory stimuli, related to Fig. 1.
a-c, Relative mRNA expression levels of pan-reactive (a), neurotoxic (b), and neuroprotective (c) substate markers in MD-astrocytes treated with PBS or ITC for 7 days. d, Heatmap depicting the expression patterns of pan-reactive astrocyte markers, neurotoxic substate markers, and neuroprotective substate markers in astrocytes treated with ITC for 6 h, followed by ITC withdrawal to 48 h. e, Heatmap depicting the expression patterns of pan-reactive astrocyte markers, neurotoxic substate markers, and neuroprotective substate markers in astrocytes treated with ITC for 2 h, followed by ITC withdrawal to 48 h. All data were presented as mean ± SEM, with n = 3 replicates per group. *P < 0.05, **P < 0.01, ***P < 0.001. In a-c, unpaired two-tailed Student’s t-tests were used. In d and e, one-way ANOVA tests were used. For each gene, the expression level was compared to that in the ITC 0 h group (*P < 0.05, an asterisk denotes significance for the average of all three replicates). The exact P values are provided in the Source data.
Extended Data Fig. 2 Occurrence of neuroprotective-to-neurotoxic substate transition in astrocytes with low-dose ITC stimulation, related to Fig. 1.
a-d, QPCR results showing the expression levels of astrocyte substates markers in MD-astrocytes stimulated with different dilutions of ITC, n = 3 replicates per group. For each gene, the expression level was compared to that in the ITC 0 h group, *P < 0.05 (an asterisk denotes significance for the average of all three replicates). One-way ANOVA tests were used for statistical analyses; the exact P values are provided in the Source data.
Extended Data Fig. 3 scRNA-seq of MD-astrocytes treated with ITC, related to Fig. 1.
a, UMAP representation displaying seven distinct clusters. b, Violin plots illustrating the expression of typical markers for astrocytes (Aqp4), glial progenitor cells (Ezh2), fibroblasts (Clo1a1), and microglia (C1qa). c, UMAP and t-SNE plots showing the classification of all cell types. d-f, Single-cell RNA-seq analysis of the expression of each marker of pan-reactive (d), neurotoxic (e), and neuroprotective (f) astrocyte substates. g, Distribution of each astrocyte subcluster along the transition trajectory.
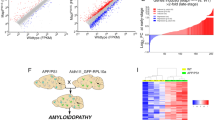
Extended Data Fig. 4 Proteomic analysis of reactive astrocyte substate transitions, related to Fig. 2.
a, Venn diagram showing the number of detected proteins in the analyzed samples. b, Scatter plot visualizing differentially expressed proteins (DEPs) observed among the four groups. c, Functional categorization of DEPs identified through proteomic analysis, classified according to the COG database. d, Heatmap displaying the differentially expressed proteins between the four groups.
Extended Data Fig. 5 Confirmation of astrocyte substate transitions in vivo, related to Fig. 3.
a, Heatmap displaying the expression patterns of neuroprotective and neurotoxic substate markers in purified brain cortical astrocytes from a mouse model injected intraperitoneally with PBS or LPS at 3- and 24-hours post-injection, n = 4 mice for saline-3h and LPS-24h groups, n = 5 mice for saline-24h and LPS-3h groups. b, Heatmap showing the expression trends of neuroprotective and neurotoxic substate markers in the whole cortical tissues of mice injected with LPS over time, n = 3 mice per group. c,d, QPCR analysis for the expression of neurotoxic (c) and neuroprotective (d) substate markers in sorted TNFRSF12A-positive astrocytes, purified from LPS-injected mice, after a 36-hour ex vivo culture in serum-free medium, n = 3 replicates per group. TNFRSF12A-negative astrocytes purified from PBS-injected mice served as control. For each marker, the mRNA expression level was normalized to that in one of the control groups. In a and b, one-way ANOVA tests were used. For each gene, the expression level was compared to that in the saline 3 h group (a) or LPS 0 h group (b), *P < 0.05 (an asterisk denotes significance for the average of all replicates). The data in c and d were presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Unpaired two-tailed Student’s t-tests were used for statistical analyses; the exact P values are provided in the Source data.
Extended Data Fig. 6 Imbalanced ratios of reactive astrocyte substates in the progression of AD and aging, related to Fig. 4.
a, Venn Diagrams showing the dynamic fluctuation balance between neurotoxic and neuroprotective substate astrocytes in 5XFAD model. The numbers in the circles indicated the cell counts of astrocytes in the neurotoxic and neuroprotective substates. For each panel, the areas of circles were proportional to the cell numbers. b, QPCR results showing the expression levels of typical markers for astrocyte substates in human astrocytes stimulated with ITC, n = 3 replicates per group. For each gene, the expression level was compared to that in the ITC 0 h group, *P < 0.05 (an asterisk denotes significance for the average of all three replicates), one-way ANOVA test was used. c, Venn Diagrams showing the dynamic fluctuation balance between neurotoxic and neuroprotective substate astrocytes in human AD. The numbers in the circles indicated the cell counts of astrocytes in the neurotoxic and neuroprotective substates. For each panel, the areas of circles were proportional to the cell numbers. d, Immunofluorescence staining showing the expression levels of neurotoxic marker C3 and neuroprotective marker PTGS2 proteins in astrocytes in the hippocampus of 3-, 6-, 12-, 18-, and 24-month-old wild-type mice (representing normal aging). Scale bar, 20 μm. e,f, Quantification of the expression levels of C3 and PTGS2 in astrocytes in d, n = 3 mice per group. The fold changes of fluorescent intensity were calculated relative to the 3-month-old group. g, Venn Diagrams showing the dynamic fluctuation balance between neurotoxic and neuroprotective substate astrocytes in normal aging condition. The numbers in the circles indicated the cell counts of astrocytes in the neurotoxic and neuroprotective substates. For each panel, the areas of circles were proportional to the cell numbers. In a, c, and g, the P values were calculated by Fisher’s exact test and showed between two groups. In e and f, data were presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA and Tukey’s multiple comparisons test were used; the exact P values are provided in the Source data.
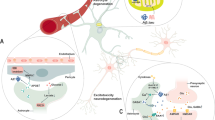
Extended Data Fig. 7 Effects of mTOR inhibitor and agonist on the expression of astrocyte substate markers, related to Fig. 5.
a, Analysis of the enrichment scores for mTOR signaling pathways in astrocytes isolated from WT and APP/PS1 mice with different ages26. The differentially expressed genes, which are the same to the genes used in Fig. 5d, in the mTOR signaling pathways were used to analyze the enrichment scores, n = 3 mice per group. b-d, Immunoblotting analysis showing the phosphorylation and expression trends of mTOR, S6, and 4EBP1 over time following IL-1α (b), TNF (c), or C1q (d) stimulation. e-h, QPCR results for the expression levels of neurotoxic substate markers of H2-T23 (e), H2-D1 (f), Gbp2 (g), and C3 (h) in astrocytes treated with saline, mTOR activator MHY1485, ITC, or the combination of MHY1485 and ITC. In e-h, data were presented as mean ± SEM, n = 3 replicates per group. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA and Tukey’s multiple comparisons test were used for each time point; the exact P values are provided in the Source data.
Extended Data Fig. 8 Effects of Rapamycin pretreatment on the expression of neurotoxic and neuroprotective substate markers, related to Fig. 6.
a,b, QPCR results showing the expression levels of neurotoxic (a) and neuroprotective (b) substate markers in MD-astrocytes pretreated with Rapamycin for 24 h and then stimulated with ITC for 24 h, n = 3 replicates per group. c, Representative images showing the expression level of neuroprotective substate marker S100A10 in SNc astrocytes after repeated Rapamycin treatment for 1 day or 5 days. d, Quantification of the expression level of S100A10 in GFAP-positive astrocytes. The fold changes of S100A10 fluorescent intensity were calculated relative to the solvent group at day 1, n = 3 mice per group. All data were presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Unpaired two-tailed Student’s t-tests were used for statistical analyses; the exact P values are provided in the Source data.
Extended Data Fig. 9 Effects of Rapamycin treatment on the expression of astrocyte substate markers in LPS-induced PD-like model, related to Fig. 6.
a, Representative images showing the expression level of neuroprotective substate marker S100A10 in SNc astrocytes. Scale bar, 50 μm. b, Quantification of the expression level of S100A10 in S100β-positive astrocytes. The fold changes of fluorescent intensity were calculated relative to the untreated control group. c, Representative images showing the expression of GFAP in the SNc. Scale bar, 100 μm. d, Quantification of the expression levels of GFAP in c. The fold changes were calculated relative to the untreated control group. All data were presented as mean ± SEM, n = 4 mice per group. *P < 0.05, ***P < 0.001. One-way ANOVA and Tukey’s multiple comparisons test were used for statistical analyses; the exact P values are provided in the Source data.
Extended Data Fig. 10 AAV-mediated knockdown of mTOR in astrocytes, related to Fig. 6.
a, Representative images showing the colocalization of mCherry with the astrocyte marker GFAP, the dopaminergic neuron marker tyrosine hydroxylase (TH), or the microglia marker Iba1 in the SNc. Scale bar, 100 μm. b, Statistical analysis of the ratio of GFAP+ mCherry+, TH+ mCherry+ and Iba1+ mCherry+ double-positive cells, indicating that mCherry was specifically expressed by astrocytes in a. c, Representative images showing the protein levels of mTOR in SNc mCherry+ GFAP+ double-positive cells of mice injected with AAVs and LPS. Scale bar, 20 μm. d, Quantification of the expression level of mTOR in astrocytes. The fold changes of mTOR fluorescent intensity were calculated relative to that in AAV-shNT group. e, Representative images showing the protein levels of neuroprotective substate marker S100A10 in SNc mCherry+ GFAP+ double-positive cells of mice injected with AAVs and LPS. Scale bar, 20 μm. f, Quantification of the expression level of S100A10 in astrocytes. The fold changes of S100A10 fluorescent intensity were calculated relative to that in AAV-shNT + LPS group. All data were presented as mean ± SEM, n = 4 mice per group. ***P < 0.001. Unpaired two-tailed Student’s t-tests were used for statistical analyses; the exact P values are provided in the Source data.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Table 1
List of identified proteins in the proteomic analysis.
Supplementary Table 2
List of DEPs in the proteomic analysis.
Supplementary Table 3
COG functional categories for DEPs.
Supplementary Table 4
List of proteins in different clusters.
Supplementary Table 5
Oligonucleotides used in this study.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Xu, Z., Jia, Z. et al. Modulating mTOR-dependent astrocyte substate transitions to alleviate neurodegeneration. Nat Aging 5, 468–485 (2025). https://doi.org/10.1038/s43587-024-00792-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-024-00792-z
This article is cited by
-
Promoting astrocyte-neuron triiodothyronine shuttling attenuates brain damage in neonatal hypoxic-ischemic encephalopathy
Journal of Neuroinflammation (2025)
-
β2 integrin regulates neutrophil trans endothelial migration following traumatic brain injury
Cell Communication and Signaling (2025)
-
Astrocyte diversity and subtypes: aligning transcriptomics with multimodal perspectives
EMBO Reports (2025)
-
Astrocytes Control Norepinephrine Signaling in the Brain
Neuroscience Bulletin (2025)