Abstract
Lung aging is a multifactorial series of molecular alterations that leads to gradual deterioration of lung function and increased vulnerability to cancer. Tumor communicates with host organs partially through extracellular vesicles; however, the mechanistic drivers and consequences of lung aging in the context of cancer remain unclear. Here we identify cancer cell-secreted dimethylarginine dimethylaminohydrolase-1 (DDAH1) protein induces citrulline accumulation and promotes lung fibrosis and aging. Mechanistically, our single-cell sequencing and genetic knockout mice evidence that citrulline availability elevation inhibits peptidyl arginine deiminase 4-mediated transforming growth factor-β1 (TGF-β1) citrullination, thereby inducing the TGF-β1/Smad3 signaling pathway in lung fibroblasts. Notably, vacuolar protein sorting assists the packaging of DDAH1 into the late endosomes. The administration of DDAH1 inhibitor reduces fibrosis and alleviates lung aging. Conclusively, our findings reveal tumor-derived DDAH1 protein contributes to citrulline accumulation to promote lung aging, shedding light on the treatment and diagnosis of tumors by inhibiting senescent lung fibroblasts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE287478 and GSE283537. The scRNA-seq data generated in this study have been deposited in the GEO under accession code GSE288993. The EV and intracellular DDAH1-interacting proteomics data measured by LC–MS-MS have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository with Project lDs IPX0008672001, IPX0010311001 and IPX0011204000, and via the Proteomics ldentifications Database partner repository with the dataset identifiers PXD069130, PXD069253 and PXD069184. The targeted LC–MS/MS data for citrulline analysis have been deposited in figshare at https://figshare.com/projects/_b_Cancer-cell-secreted_DDAH1_induces_TGF-_1_Smad3_signaling_pathway_to_promote_fibrosis_and_aging_in_lung_b_/260780. The mass spectrometry proteomics data reported in this paper are provided in Supplementary Table 3 and Supplementary Table 4. Source data containing the graphical representations, statistical descriptions and uncropped gels and blots are provided with the paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Kroemer, G. et al. From geroscience to precision geromedicine: Understanding and managing aging. Cell 188, 2043–2062 (2025).
Tabula Muris, C. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Kaeberlein, M., Rabinovitch, P. S. & Martin, G. M. Healthy aging: the ultimate preventative medicine. Science 350, 1191–1193 (2015).
Calhoun, C. et al. Senescent cells contribute to the physiological remodeling of aged lungs. J. Gerontol. A Biol. Sci. Med. Sci. 71, 153–160 (2016).
Yanai, H. et al. Cellular senescence-like features of lung fibroblasts derived from idiopathic pulmonary fibrosis patients. Aging 7, 664–672 (2015).
Yang, K. E. et al. Differential expression of extracellular matrix proteins in senescent and young human fibroblasts: a comparative proteomics and microarray study. Mol. Cells 32, 99–106 (2011).
Fane, M. E. et al. Stromal changes in the aged lung induce an emergence from melanoma dormancy. Nature 606, 396–405 (2022).
Meguro, S. et al. Preexisting senescent fibroblasts in the aged bladder create a tumor-permissive niche through CXCL12 secretion. Nat. Aging 4, 1582–1597 (2024).
Budinger, G. R. S. et al. The Intersection of Aging Biology and the Pathobiology of Lung Diseases: A Joint NHLBI/NIA Workshop. J. Gerontol. A Biol. Sci. Med. Sci. 72, 1492–1500 (2017).
Lu, Z. et al. Epigenetic therapy inhibits metastases by disrupting premetastatic niches. Nature 579, 284–290 (2020).
Li, Z. et al. Tumor-produced and aging-associated oncometabolite methylmalonic acid promotes cancer-associated fibroblast activation to drive metastatic progression. Nat. Commun. 13, 6239 (2022).
Patel, U., Susman, D. & Allan, A. L. Influence of extracellular vesicles on lung stromal cells during breast cancer metastasis. Int. J. Mol. Sci. https://doi.org/10.3390/ijms241411801 (2023).
Becker, A. et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 30, 836–848 (2016).
Chen, X. et al. Small extracellular vesicles from young plasma reverse age-related functional declines by improving mitochondrial energy metabolism. Nat. Aging 4, 814–838 (2024).
Tarbit, E., Singh, I., Peart, J. N. & Rose’Meyer, R. B. Biomarkers for the identification of cardiac fibroblast and myofibroblast cells. Heart Fail. Rev. 24, 1–15 (2019).
Revy, P., Kannengiesser, C. & Bertuch, A. A. Genetics of human telomere biology disorders. Nat. Rev. Genet. 24, 86–108 (2023).
Ostrowski, M. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 (2010).
Takasugi, M. et al. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 8, 15729 (2017).
Borghesan, M. et al. Small extracellular vesicles are key regulators of non-cell autonomous intercellular communication in senescence via the interferon protein IFITM3. Cell Rep. 27, 3956–3971 e3956 (2019).
Kaur, A. et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 532, 250–254 (2016).
Lee, S. et al. Molecular programs of fibrotic change in aging human lung. Nat. Commun. 12, 6309 (2021).
Hoshino, A. et al. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015).
Palm, F., Onozato, M. L., Luo, Z. & Wilcox, C. S. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. Am. J. Physiol. Heart Circ. Physiol. 293, H3227–3245 (2007).
Chaleckis, R., Murakami, I., Takada, J., Kondoh, H. & Yanagida, M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl Acad. Sci. USA 113, 4252–4259 (2016).
Kouchiwa, T. et al. Age-related changes in serum amino acids concentrations in healthy individuals. Clin. Chem. Lab. Med. 50, 861–870 (2012).
Pullamsetti, S. S. et al. The role of dimethylarginine dimethylaminohydrolase in idiopathic pulmonary fibrosis. Sci. Transl. Med. 3, 87ra53 (2011).
Schneider, J. L. et al. The aging lung: physiology, disease, and immunity. Cell 184, 1990–2019 (2021).
Hong, L. & Fast, W. Inhibition of human dimethylarginine dimethylaminohydrolase-1 by S-nitroso-L-homocysteine and hydrogen peroxide. Analysis, quantification, and implications for hyperhomocysteinemia. J. Biol. Chem. 282, 34684–34692 (2007).
Hannemann, J. et al. L-arginine dependence of breast cancer - molecular subtypes matter. BMC Cancer https://doi.org/10.1186/s12885-025-13908-4 (2025).
Lee, Y. C. et al. L-Arginine and L-citrulline supplementation have different programming effect on regulatory T-cells function of infantile rats. Front. Immunol. 9, 2911 (2018).
Ren, L. L. et al. TGFβ as a master regulator of aging-associated tissue fibrosis. Aging Dis. 14, 1633–1650 (2023).
Yuzhalin, A. E. Citrullination in cancer. Cancer Res. 79, 1274–1284 (2019).
Sipila, K. H. et al. Extracellular citrullination inhibits the function of matrix associated TGF-β. Matrix Biol. 55, 77–89 (2016).
Mondal, S. & Thompson, P. R. Protein arginine deiminases (PADs): biochemistry and chemical biology of protein citrullination. Acc. Chem. Res. 52, 818–832 (2019).
Tan, W. et al. Hsp90 Inhibitor STA9090 induced VPS35 related extracellular vesicle release and metastasis in hepatocellular carcinoma. Transl. Oncol. 26, 101502 (2022).
Ghebremariam, Y. T., Erlanson, D. A. & Cooke, J. P. A novel and potent inhibitor of dimethylarginine dimethylaminohydrolase: a modulator of cardiovascular nitric oxide. J. Pharmacol. Exp. Ther. 348, 69–76 (2014).
Belle, J. I. et al. Senescence defines a distinct subset of myofibroblasts that orchestrates immunosuppression in pancreatic cancer. Cancer Discov. 14, 1324–1355 (2024).
Ye, J. et al. Senescent CAFs mediate immunosuppression and drive breast cancer progression. Cancer Discov. 14, 1302–1323 (2024).
Fane, M. & Weeraratna, A. T. How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 20, 89–106 (2020).
Fan, L. et al. PADI4 epigenetically suppresses p21 transcription and inhibits cell apoptosis in fibroblast-like synoviocytes from rheumatoid arthritis patients. Int J. Biol. Sci. 13, 358–366 (2017).
Ragavan, V. N. et al. A multicentric consortium study demonstrates that dimethylarginine dimethylaminohydrolase 2 is not a dimethylarginine dimethylaminohydrolase. Nat. Commun. 14, 3392 (2023).
Li, M. X. et al. Tumor-derived miR-9-5p-loaded EVs regulate cholesterol homeostasis to promote breast cancer liver metastasis in mice. Nat. Commun. 15, 10539 (2024).
Guan, R. et al. Bone morphogenetic protein 4 inhibits pulmonary fibrosis by modulating cellular senescence and mitophagy in lung fibroblasts. Eur. Respir. J. https://doi.org/10.1183/13993003.02307-2021 (2022).
Seluanov, A., Vaidya, A. & Gorbunova, V. Establishing primary adult fibroblast cultures from rodents. J. Vis. Exp. https://doi.org/10.3791/2033 (2010).
Bi, Y. K. et al. Exosomal miR-302b rejuvenates aging mice by reversing the proliferative arrest of senescent cells. Cell Metab. https://doi.org/10.1016/j.cmet.2024.11.013 (2025).
Katsumura, S. et al. Deadenylase-dependent mRNA decay of GDF15 and FGF21 orchestrates food intake and energy expenditure. Cell Metab. 34, 564–580 e568 (2022).
Wang, C. et al. Leukocyte telomere length in children born following blastocyst-stage embryo transfer. Nat. Med. 28, 2646–2653 (2022).
Zhang, X. et al. Arachidonic acid triggers spermidine synthase secretion from primary tumor to induce skeletal muscle weakness upon irradiation. Cell Metab. 37, 1766–1782 e1712 (2025).
Jiang, B. et al. Progerin modulates the IGF-1R/Akt signaling involved in aging. Sci. Adv. 8, eabo0322 (2022).
Xiao, J. et al. 25-Hydroxycholesterol regulates lysosome AMP kinase activation and metabolic reprogramming to educate immunosuppressive macrophages. Immunity 57, 1087–1104 e1087 (2024).
Liu, Y. et al. Artemisinins ameliorate polycystic ovarian syndrome by mediating LONP1-CYP11A1 interaction. Science 384, eadk5382 (2024).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Zheng, L. T. et al. Pan-cancer single cell landscape of tumor-infiltrating T cells. Science 374, 1462 (2021).
Abdullah, M. et al. Cholesterol regulates exosome release in cultured astrocytes. Front. Immunol. 12, 722581 (2021).
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2021YFA0804803), National Natural Science Foundation of China to W.Y. (32270827, 82203590) and Fundamental Research Funds for the Central Universities (2042025kf0003). We thank all the core facilities (including Mass Spectrometry, Microscopy Core, Electron Microscopy facility and Animal Core) of the College of Life Sciences, Ultracentrifugation Platform provided by the State Key Laboratory of Virology and the Large Instruments and Equipment Open Sharing Subsidies of Wuhan University. We are grateful to B. Wu and Y. Liu from Wuhan University for their assistance in single-cell sequencing analysis and providing the aged mice, respectively.
Author information
Authors and Affiliations
Contributions
W.Y.: conceptualization, methodology, supervision, funding acquisition, project administration and writing-review and editing. L.L.: methodology, visualization, investigation, formal analysis, writing-review and editing and figure generating. J.L., A.C. and Z.Y.: resources. Q.W., M.C. and H.Z.: software and formal analysis. X.L., Y.H., C.W., M.L., S.H., X.Z. and X.W.: methodology and investigation. All authors read and approved the manuscript. This statement is based on the Contributor Roles Taxonomy (CRediT).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Tohru Minamino and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
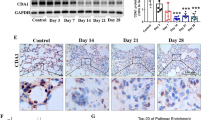
Extended Data Fig. 1 Breast cancer-derived extracellular vesicles promote lung aging.
a, Western blot showing RAB27A protein level in 4T1/Rab27a KO cells; β-Actin served as a loading control for immunoblot. Repeated three times independently with similar results obtained. b, Particles number of EVs derived from 4T1/Rab27a KO cells compared to 4T1/Ctrl cells (n = 3 biologically independent samples per group). c, Experimental diagram for the tumor-bearing mouse model. Mice were randomly divided into three groups, the tumor-free group and the other two groups injected with 4T1/Ctrl or 4T1/Rab27a KO tumor cells labeled lck-GFP under the fourth pair of mammary fat pads respectively. d, Fluorescence signals in lungs 5 weeks after subcutaneous seeding of Lck-GFP-labeled 4T1/Ctrl or 4T1/Rab27a KO tumor cells. e, GFP signals (green) in lung tissues from the mice bearing Lck-GFP labeled tumor cells. Repeated three times independently with similar results obtained. Scale bar, 200 μm. f, Tumor volumes were measured every 2 days (n = 9 mice per group). g, Body weight was measured every 2 days (n = 9 mice per group). h, Representative images and corresponding quantitative analysis of senescence-associated β-galactosidase staining (Scale bar, 50 μm; n = 7 mice per group). Arrows indicated regions that were positive for β-galactosidase staining. i, Representative p21 immunohistochemistry (IHC) staining images and corresponding quantitative analysis of lung tissues from mice in tumor-bearing group (Scale bar, 100 μm; n = 7 mice per group). Quantification was performed by using ImageJ software, with integrated optical density (IOD) values normalized to the tumor-free group. j, Representative H&E staining images and corresponding quantitative analysis of lung tissues from tumor-bearing group mice (Scale bar, 200 μm; n = 10 mice per group). k, Representative Sirius red staining images and corresponding quantitative analysis of lung tissues from tumor-bearing group mice (Scale bar, 200 μm; n = 8 mice per group). l, Representative α-SMA IHC staining images and corresponding quantitative analysis of the lung tissues from mice of indicated groups (Scale bar, 200 μm; n = 5 mice per group). Quantification was normalized to tumor-free group. m, Standardized telomere length of lungs from mice in the indicated groups (n = 5 mice per group). In the natural aging group, values were normalized to young mice; in the tumor-bearing group, values were normalized to tumor-free mice. When aged mice were used as the reference, comparative analyses were performed between the aged mice and the tumor-bearing group mice. The center line represents the median, box edges delineate first and third quartiles and whiskers extend to minimum and maximum values. BioRender (https://biorender.com/) was used for c. Data were presented as mean ± s.e.m. (Standard Error of the Mean); exact P values were shown and reported as source data; unpaired two-tailed Student’s t-test was used for b; two-way analysis of variance (ANOVA) was used for f, g; one-way ANOVA followed by Dunnett multiple comparison test was used for h, i, j, k, l, m.
Extended Data Fig. 2 Tumor-derived exosomal DDAH1 promotes aging in lung fibroblasts.
a-b, DDAH1 protein level in 10A/DDAH1, 231/DDAH1 KO, 4T1/DDAH1 KO cells, and indicated EVs; β-Actin (a cytoplasmic control) or Syntenin-1 (an EVs control) served as a loading control. c, 10A/DDAH1, 231/DDAH1 KO, and 4T1/DDAH1 KO EV fractions showing both positive markers and negative marker (GM130) of EVs in OptiPrep gradient fractions. Intensity of blots was quantified with ImageJ (quantification shown below images). d, Western blot showing levels of indicated proteins in lung tissues from tumor-bearing groups of mice; β-Actin served as a loading control. e, Representative IHC staining images and corresponding quantitative analysis of DDAH1 in tumor-bearing mice lung tissues (Scale bar, 200 μm; n = 7 mice per group). Quantification was normalized to 4T1/Ctrl tumor-bearing group mice. f, Standardized telomere length of lungs from mice in the indicated groups (n = 6 mice per group, values were normalized to 4T1/Ctrl tumor-bearing group). The center line represents the median, box edges delineate first and third quartiles and whiskers extend to minimum and maximum values. g, Body weight and tumor volumes were measured every 2 days (n = 6 mice per group). h, Bioluminescence imaging of lung tissues dissected from 4T1/Ctrl and 4T1/DDAH1 KO tumor-bearing mice. Luminescence signals were captured via the IVIS Spectrum system and color-coded on intensity. i, Representative H&E staining images and corresponding quantitative analysis of 4T1/Ctrl and 4T1/DDAH1 KO mice lung metastasis (Scale bar, 1 mm; n = 5 mice per group). Quantification was performed with ImageScope software. j, Fraction of ten major classes of cells were detected in the lungs of tumor-bearing mice samples. k, Volcano plots based on single-cell differential expression analysis of lung fibroblasts from 4T1/Ctrl group comparing with 4T1/DDAH1 KO group mice. Genes related to aging were highlighted. l, UMAP visualization showing the subtypes of fibroblast clusters. Bar plots showing the fraction of fibroblast clusters. m, Heatmap showing the expression pattern of notably highly expressed genes for each fibroblast clusters, with top marker genes highlighted. Z score normalized mean expression. n, Bar plots showing the proportion of young fibroblast and senescence-like fibroblast in total fibroblasts and in each fibroblast cluster. o, Representative immunofluorescence staining images and corresponding quantitative analysis of the colocalization of DDAH1 and α-SMA in lung tissues from 10A/Ctrl and 10A/DDAH1 EV injection mice (Scale bar, 100 μm; n = 3 mice per group). p, MPFs isolated from mice lungs were identified by immunofluorescence staining for vimentin in green fluorescence and DAPI in blue. Scale bar, 20 μm. The lower panel displaying GFP signal of MPF treated with Lck-GFP-labeled 231 EVs. Scale bar, 20 μm. Experiments were repeated three times for a-d. Representative images are displayed. Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data; unpaired two-tailed Student’s t-test was used for e, f, i, o; two-way ANOVA was used for g; two- tailed Wilcoxon rank-sum test was used for k.
Extended Data Fig. 3 DDAH1 induces citrulline accumulation to promote lung aging.
a, Measurement of DDAH1 activity in indicated EVs, with colorimetric detection to determine citrulline level produced by equal amounts of indicated EVs (n = 3 independent replicates). b, Schematic diagram depicted the sites of enzymatically inactive mutant in the DDAH1. c, Western blot showing the levels of indicated proteins in WI-38 cells transfected with wild-type DDAH1 and the enzymatically mutated DDAH1(D127A/H173A/C274A); β-Actin served as a loading control. Repeated three times independently with similar results obtained. d, GFP signals (green) in indicated tissues of AAV6 intranasal administration model mice, including lung, brain, liver, kidney, spleen and heart. Scale bar, 200 μm. e, Representative IHC staining images and the quantification of images of DDAH1 and α-SMA in lung tissues from AAV6 intranasal administration model mice (Scale bar, 200 μm; n = 5 mice per group). Quantification was normalized to AAV-GFP group mice. f, Real-time qPCR analysis of Cdkn1a, Cdkn2a, Acta2, Col1a1, Col3a1, Ddr1, Fgf2, Ctgf, Il1b, Il6, Mmp9, and Mmp12 mRNA abundance in lung tissues from AAV6 intranasal administration group mice (n = 6 mice per group), values were normalized to AAV-GFP group mice. g, Representative Sirius red staining images and corresponding quantitative analysis of lung tissues in mice from AAV intranasal administration model (Scale bar, 200 μm; n = 8 mice per group). h, Representative H&E staining images and corresponding quantitative analysis of lung tissues from AAV intranasal administration group mice (Scale bar, 200 μm; n = 10 mice per group). i, Real-time qPCR analysis of Cdkn1a, Cdkn2a, Acta2, Col1a1, Col3a1, Ddr1, Fgf2, Ctgf, Il1b, Il6, Mmp9, and Mmp12 mRNA abundance in MPFs isolated from indicated group mice (n = 3 biologically independent samples per group). Values were normalized to MPFs isolated from AAV-GFP group mice lungs. j, Population doubling curves of indicated MPFs; n = 3 biologically independent samples per group. k, Representative senescence-associated β-galactosidase staining images and corresponding quantitative analysis of MPFs following 10 days exposure to 20 μM citrulline (Scale bar, 100 μm; n = 10 biologically independent samples per group). l, Population doubling curves of MPFs exposure to 20 μM citrulline or PBS (solvent control); n = 3 biologically independent samples per group. m, Real-time qPCR analysis of Cdkn1a, Cdkn2a, Acta2, Col1a1, Col3a1, Ddr1, Fgf2, Ctgf, Il1b, Il6, Mmp9, and Mmp12 mRNA abundance in lung tissues from citrulline intranasal administration model group mice (n = 8 mice per group). Values were normalized to PBS intranasal administration group mice. n, Representative H&E staining images and corresponding quantitative analysis of lung tissues in mice from PBS/citrulline intranasal administration model (Scale bar, 200 μm; n = 8 mice per group). o, Representative images of Sirius red staining and corresponding quantitative analysis with lung tissues from PBS or citrulline intranasal administration model mice (Scale bar, 200 μm; n = 8 mice per group). p, Representative images and corresponding quantitative analysis of senescence-associated β-galactosidase staining (Scale bar, 50 μm; n = 8 mice per group). The area of SA-β-gal-positive staining was quantified using ImageJ software. Arrows indicated regions that were positive for β-galactosidase staining. q, Representative Sirius red staining images and corresponding quantitative analysis of lung tissues in mice from citrulline intranasal administration tumor-bearing mouse model (Scale bar, 200 μm; n = 8 mice per group). Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data; unpaired two-tailed Student’s t-test was used for a, k, m, n, o, p, q; one-way ANOVA followed by Dunnett multiple comparison test was used for e, f, g, h, i; two-way ANOVA was used for j, l.
Extended Data Fig. 4 Citrulline promotes TGF-β1/SMAD3 signaling pathway in lung fibroblasts.
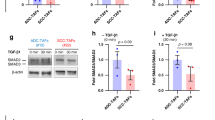
a, Western blot analysis of indicated proteins in MPFs treated with indicated EVs or PBS; β-Actin served as a loading control for the immunoblot. Repeated three times independently with similar results obtained. b, Western blot analysis of t-SMAD3 (total SMAD3) and p-SMAD3 (phosphorylated of SMAD3 at S423/S425) in nucleus and cytoplasmic fractions of WI-38 cells treated with indicated EVs. H3 (a nuclear control) or β-Actin (a cytoplasmic control) served as a loading control. Blots were quantified with ImageJ (quantification of proteins in nucleus or cytoplasmic fractions normalized to respective loading control and shown below image). Experiments were repeated three times independently with consistent results. c, Representative SMAD3 immunofluorescence images and corresponding quantitative analysis in MPFs treated with indicated EVs or PBS. Scale bar, 10 μm; n = 15 cells per group. d, Representative SMAD3 immunofluorescence images and corresponding quantitative analysis in MPFs following 24-hour exposure to 20 μM citrulline or PBS (Scale bar, 10 μm; n = 15 cells per group). e, Western blot analysis of t-SMAD3 (total SMAD3) and p-SMAD3 (phosphorylated of SMAD3 at S423/S425) in nucleus and cytoplasmic fractions of MPFs exposure to 20 μM citrulline or PBS (solvent control). H3 or β-Actin served as a loading control. Blots were quantified with ImageJ (quantification of proteins in nucleus or cytoplasmic fractions normalized to respective loading control and shown below image). Experiments were repeated three times independently with consistent results. f, Stability of the indicated mRNAs in MPFs treated with 20 μM citrulline or PBS (solvent control) incubated with 5 μg/mL of Actinomycin D for the indicated time; n = 3 biologically independent samples per group. g-h, Real-time qPCR analysis of Tgfb1, Tgfb2 and Tgfb3 mRNA abundance in lung tissues from indicated EV tail vein injection group and tumor-bearing group mice (n = 7 mice per group). i, Western blot showing the TGF-β2 and TGF-β3 protein levels of the lungs from indicated group mice; β-Actin served as a loading control. Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data. One-way ANOVA followed by Dunnett multiple comparison test was used for c, g, h; unpaired two-tailed Student’s t-test was used for d; two-way ANOVA was used for f.
Extended Data Fig. 5 Citrulline inhibits PAD4-mediated TGF-β1 citrullination.
a, Western blot showing the citrullination level of TGF-β1 in MPFs treated with PBS or indicated EVs, total TGF-β1 level served as a loading control. b, Real-time qPCR analysis of Padi2 mRNA abundance in WI-38/PADI2 KD cells compared with WI-38 cells transfected with plko.1 vector; n = 3 biologically independent samples per group. Values were normalized to vector control. Data were presented as mean ± s.e.m. c, Real-time qPCR analysis of Padi4 mRNA abundance in WI-38/PADI4 KD cells compared with WI-38 cells transfected with plko.1 vector; n = 3 biologically independent samples per group. Values were normalized to vector control. Data were presented as mean ± s.e.m. d-e, Western blot showing indicated protein levels in WI-38/PADI2 KD cells and WI-38/PADI4 KD cells following 48-hour exposure to 20 μM citrulline or PBS (solvent control) compared with WI-38 cells transfected with plko.1 vector; β-Actin served as a loading control. f-g, Western blot showing the citrullination level of TGF-β1 in WI-38 cells with PAD2 or PAD4 knockdown (WI-38/PADI2 KD or WI-38/PADI4 KD) following 24-hour exposure to 20 μM citrulline or PBS (solvent control) compared to plko.1 vector-transfected controls. Total TGF-β1 level was used as a loading control. h, Genotype identification of Padi4−/− mice. i, MPFs treated with PBS, indicated EVs or 20 μM citrulline were subjected to SMAD3 immunofluorescence staining and confocal microscopic imaging. Representative images were shown. Scale bar, 10 μm. j, MPFs from wild-type C57BL/6 J mice or Padi4−/− mice were treated with 20 μM citrulline or PBS to detect the amounts of t-SMAD3 (total SMAD3), p-SMAD3 (phosphorylated of SMAD3 at S423/S425) in the nucleus and cytoplasmic fractions by western blot. H3 (a nuclear control) or β-Actin (a cytoplasmic control) served as a loading control. Blots were quantified with ImageJ (quantification of proteins in nucleus or cytoplasmic fractions normalized to respective loading control and shown below image). k, Western blot showing the citrullination level of TGF-β1 in wild-type and Padi4−/− MPFs following 24-hour exposure to 20 μM citrulline or PBS (solvent control), total TGF-β1 level served as a loading control. l, Western blot showing the citrullination level of H2A, H3, H4, and GSK3β in wild-type and Padi4−/− MPFs exposure to 20 μM citrulline or PBS (solvent control), total indicated protein level served as a loading control. Blots were quantified with ImageJ (values under each blot were normalized to the corresponding control and subsequent to the first lane). m, Population doubling curves of indicated MPFs isolated from Padi4−/− mice; n = 3 biologically independent samples per group. n, Representative H&E staining images and corresponding quantitative analysis of lung tissues in Padi4−/− C57 mice from PBS/citrulline intranasal administration model (Scale bar, 200 μm; n = 8 mice per group). Data were presented as mean ± s.e.m. o, Representative Sirius red staining images and corresponding quantitative analysis of lung tissues from Padi4−/− C57 mice with PBS/citrulline intranasal administration (Scale bar, 100 μm; n = 7 mice per group). Data were presented as mean ± s.e.m. p, Representative IHC staining images and corresponding quantitative analysis of DDAH1 in indicated EV injection group Padi4−/− mice lung tissues (Scale bar, 100 μm; n = 7 mice per group). Data were presented as mean ± s.e.m. q, Representative images and corresponding quantitative analysis of H&E staining (Scale bar, 100 μm; n = 7 mice per group). Data were presented as mean ± s.e.m. r, Representative Sirius red staining images and corresponding quantitative analysis of lung tissue in indicated EV injection group Padi4−/− mice (Scale bar, 200 μm; n = 7 mice per group). Data were presented as mean ± s.e.m. s, Western blot showing the citrullination level of TGF-β1 in WI-38 cells transfected with wild-type PAD4 or the E411A and R651A double mutated PAD4 following 24-hour exposure to 20 μM citrulline or PBS, total TGF-β1 level served as a loading control. Experiments were repeated three times for a, d-g, j-l, s. Representative images are displayed. Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data. One-way ANOVA followed by Dunnett multiple comparison test was used for b, c, p, q, r; two-way ANOVA was used for m; unpaired two-tailed Student’s t-test was used for n, o.
Extended Data Fig. 6 VPS35 assists DDAH1 secretion and integrin facilitates DDAH1 lung targeting.
a, Western blot showing the protein level of VPS35 in whole-cell lysates of 4T1/VPS35 KD cells compared with 4T1/Ctrl cells; β-Actin served as a loading control. Repeated three times independently with similar results obtained. b, DDAH1 protein level in 4T1/VPS35 KD EVs; Syntenin-1 served as a loading control. Repeated three times independently with similar results obtained. c, Representative mIHC staining images and quantitative analysis of DDAH1 and CD63 in 4T1/Ctrl or 4T1/VPS35 KD tumors (Scale bar, 20 μm; n = 3 mice per group). d, DDAH1 protein level in circulating EVs extracted from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing mice serum, ALIX served as a loading control. e, Representative IHC staining images and corresponding quantitative analysis of DDAH1 in 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing mice lung tissues (Scale bar, 100 μm; n = 8 mice per group). Quantification was normalized to the 4T1/Ctrl group. f, Citrulline level in lung tissues from the indicated group of mice (n = 7 mice per group). g, Western blot showing the protein levels of the indicated proteins in lungs from tumor-bearing group mice; β-Actin served as a loading control. h, Representative IHC staining images and corresponding quantitative analysis of TGF-β1 in lung tissues from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing group mice (Scale bar, 100 μm; n = 8 mice per group). The quantification analysis was normalized to 4T1/Ctrl group. i, Representative senescence-associated β-galactosidase staining images and corresponding quantitative analysis with lung tissues from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing group mice (Scale bar, 50 μm; n = 8 mice per group). j, Representative p21 IHC staining images and corresponding quantitative analysis of lung tissues from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing group mice (Scale bar, 100 μm; n = 8 mice per group). Quantification was normalized to 4T1/Ctrl group mice. k, Representative H&E staining images and corresponding quantitative analysis of lung tissues in mice from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing group (Scale bar, 200 μm; n = 7 mice per group). l, Representative Sirius red staining images and corresponding quantitative analysis of lung tissues from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing group mice (Scale bar, 200 μm; n = 7 mice per group). m, Real-time qPCR analysis of Cdkn1a, Cdkn2a, Acta2, Col1a1, Col3a1, Ddr1, Fgf2, Ctgf, Il1b, Il6, Mmp9, Mmp12 mRNA abundance in lung tissues from 4T1/Ctrl and 4T1/VPS35 KD tumor-bearing group mice (n = 6 mice per group). Values were normalized to 4T1/Ctrl group mice. n, Proteomic Profiling of proteins in 231 EV. ITGα6 and ITGβ4 were highlighted. o, Proteomic Profiling of ITGα6 and ITGβ4 in IP–MS. p-q, Interaction between Flag-tagged DDAH1 and ITGβ4/α6 was verified by co-IP experiment and immunoblot detection. Intensity of blots was quantified with ImageJ (quantification shown below images). Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data; unpaired two-tailed Student’s t-test was used for c, e, f, h, i, j, k, l, m.
Extended Data Fig. 7 DDAH1/TGF-β1 axis positively correlates with breast cancer lung aging and metastasis.
a, Western blot showing DDAH1 protein level of the tumor and normal tissue adjacent to the tumor from human breast cancer patients (n = 4 pairs of samples; N: normal; T: tumor), β-Actin served as a loading control. b, Real-time qPCR analysis of DDAH1 mRNA abundance in indicated cells (n = 3 biologically independent samples per group). Values were normalized to MCF-10A cells. c-d, Western blot analysis of DDAH1 protein level in whole-cell lysates and EVs from indicated cells. β-Actin or Syntenin-1 served as a loading control. e, Representative mIHC staining images of α-SMA, DDAH1, PAD4, TGF-β1, and p21 in indicated group mice lung tissues. Scale bar, 100 μm. f, Representative IHC staining images (DDAH1, p21, p16, TGF-β1, Ki67), and representative Sirius red staining images of lung tissues from breast cancer patients with high level and low level of DDAH1 (Scale bar, 200 μm). g, DDAH1 expression level in breast cancer with lung metastasis and the other organ metastasis obtained from HCMDB database (n = 7 samples for lung metastasis; n = 15 samples for other organ metastasis). The center line represents the median, box edges delineate first and third quartiles and whiskers extend to minimum and maximum values. h, Western blot showing the protein level of DDAH1 in circulating EVs extracted from 24-month-old mice and 2-month-old mice serum, ALIX served as a loading control. i-j, Representative DDAH1 and TGF-β1 IHC staining images and corresponding quantitative analysis in PD 404182 and solvent-treated tumor-bearing mice lungs (Scale bar, 100 μm; n = 6 mice per group for DDAH1 IHC; n = 8 mice per group for TGF-β1 IHC). Quantification was normalized to solvent control group mice. k, Representative images of Sirius red staining and corresponding quantitative analysis of lung tissues from PD 404182 or solvent-treated tumor-bearing mice (Scale bar, 200 μm; n = 8 mice per group). l, Real-time qPCR analysis of Cdkn1a, Cdkn2a, Acta2, Col1a1, Col3a1, Ddr1, Fgf2, Ctgf, Il1b, Il6, Mmp9, and Mmp12 mRNA abundance in lung tissues from PD 404182 or solvent-treated tumor-bearing mice (n = 6 mice per group), values were normalized to solvent control group mice. m, Western blot showing the levels of indicated proteins in lung tissues from PD 404182 or solvent-treated tumor-bearing mice; β-Actin served as a loading control. Experiments were repeated three times for a, c-e. Representative images are displayed. Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data; one-way ANOVA followed by Dunnett multiple comparison test was used for b; two-tailed Mann–Whitney U-test was used for g; unpaired two-tailed Student’s t-test was used for i, j, k, l.
Extended Data Fig. 8 DDAH2 fails to induce lung aging and fibrosis.
a, DDAH2 protein level in 231/Ctrl and 231/DDAH2 KO cells, and their derived EVs; β-Actin (a cytoplasmic control) or Syntenin-1 (an EVs control) served as a loading control. Blots were quantified with ImageJ (values under each blot were normalized to the corresponding control and subsequent to the first lane). Repeated three times independently with similar results obtained. b, Experimental diagram for the tumor-bearing mouse model. Mice were randomly divided into two groups, injected with 231/Ctrl or 231/DDAH2 KO tumor cells under the fourth pair of mammary fat pads respectively. c, Citrulline level in lung tissues from the 231/Ctrl and 231/DDAH2 KO tumor-bearing group mice (n = 7 mice per group). d, Real-time qPCR analysis of Cdkn1a, Cdkn2a, Acta2, Col1a1, Col3a1, Ddr1, Fgf2, Ctgf, Il1b, Il6, Mmp9, and Mmp12 mRNA abundance in lung tissues from 231/Ctrl and 231/DDAH2 KO tumor-bearing groups (n = 6 mice per group). Values were normalized to 231/Ctrl tumor-bearing group mice. e, Western blot showing levels of indicated proteins in lung tissues from 231/Ctrl and 231/DDAH2 KO tumor-bearing groups mice; β-Actin served as a loading control. Blots were quantified with ImageJ (values under each blot were normalized to the corresponding control and subsequent to the first lane). f, Representative IHC staining images and corresponding quantitative analysis of TGF-β1 in 231/Ctrl and 231/DDAH2 KO tumor-bearing mice lung tissues (Scale bar, 100 μm; n = 7 mice per group). Quantification was performed by using ImageJ software, with integrated optical density (IOD) values normalized to 231/Ctrl tumor-bearing group mice. g, Representative images and corresponding quantitative analysis of senescence-associated β-galactosidase staining (Scale bar, 100 μm; n = 7 mice per group). The area of SA-β-gal-positive staining was quantified using ImageJ software. Arrows indicated regions that were positive for β-galactosidase staining. h, Representative H&E staining images and corresponding quantitative analysis of lung tissues from 231/Ctrl and 231/DDAH2 KO tumor-bearing group mice (Scale bar, 100 μm; n = 7 mice per group). i, Representative Sirius red staining images and corresponding quantitative analysis of lung tissues from tumor-bearing group mice (Scale bar, 200 μm; n = 7 mice per group). j, Western blot showing levels of indicated proteins in MPFs following 10 days exposure to indicated EVs or PBS (solvent control); β-Actin served as a loading control. Blots were quantified with ImageJ (values under each blot were normalized to the corresponding control and subsequent to the first lane). Repeated three times independently with similar results obtained. BioRender (https://biorender.com/) was used for b. Data were presented as mean ± s.e.m.; exact P values were shown and reported as source data; unpaired two-tailed Student’s t-test was used for c, d, f, g, h, i.
Supplementary information
Supplementary Tables 1–6
Primers used in this study; antibodies used in this study; proteins determined by quantitative proteomics for EVs; DDAH1-interacting protein candidates identified by IP–MS analysis; clinical information included in this study; and marker genes for cell type identification.
Source data
Source Data Fig. 1
Statistical source data and unprocessed western blots.
Source Data Fig. 2
Statistical source data and unprocessed western blots.
Source Data Fig. 3
Statistical source data and unprocessed western blots.
Source Data Fig. 4
Statistical source data and unprocessed western blots.
Source Data Fig. 5
Statistical source data and unprocessed western blots.
Source Data Fig. 6
Statistical source data and unprocessed western blots.
Source Data Fig. 7
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data and unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, L., Wang, Q., Chen, M. et al. Cancer-cell-secreted DDAH1 induces TGF-β1/Smad3 signaling pathway to promote fibrosis and aging in lung. Nat Aging 6, 143–161 (2026). https://doi.org/10.1038/s43587-025-01024-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-01024-8