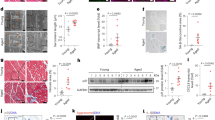
Abstract
Cardiac aging is a major driver of cardiovascular diseases and associated mortality, yet its therapeutic options are limited. While long interspersed nuclear element-1 (LINE-1) retrotransposons are known to drive cellular senescence, their role in cardiac aging is poorly defined. Here we showed that LINE-1 expression increased in the heart with age. To investigate their role in cardiac aging, we generated cardiomyocyte-specific Mov10-knockout mice, which failed to suppress LINE-1. These mice developed LINE-1 derepression, cardiac dysfunction and premature cardiac aging by 3 months of age, accompanied by cGAS–STING activation. Pharmacological inhibition of LINE-1 reverse transcription (with 3TC) or STING (with H-151) suppressed cGAS–STING activation and attenuated senescence in Mov10-knockout H9C2 cells. Notably, both inhibitors improved cardiac function and reduced cardiac inflammation and senescence phenotypes in naturally aged mice. Together, our findings establish LINE-1 as a driver of cardiac aging via cGAS–STING activation, highlighting LINE-1 and its downstream effectors as therapeutic targets for age-related cardiac dysfunction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All the RNA-sequencing data have been deposited to the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA1338093. Source data are provided with this paper.
References
Abdellatif, M., Rainer, P. P., Sedej, S. & Kroemer, G. Hallmarks of cardiovascular ageing. Nat. Rev. Cardiol. 20, 754–777 (2023).
Vakka, A., Warren, J.S. & Drosatos, K. Cardiovascular aging: from cellular and molecular changes to therapeutic interventions. J. Cardiovasc. Aging 3, 23 (2023).
Li, X. et al. LINE-1 transcription activates long-range gene expression. Nat. Genet. 56, 1494–1502 (2024).
Beck, C. R., Garcia-Perez, J. L., Badge, R. M. & Moran, J. V. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 12, 187–215 (2011).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Tang, H. et al. The transcription factor PAX5 activates human LINE1 retrotransposons to induce cellular senescence. EMBO Rep. 25, 3263–3275 (2024).
Simon, M. et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885 (2019).
Arjan-Odedra, S., Swanson, C. M., Sherer, N. M., Wolinsky, S. M. & Malim, M. H. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 9, 53 (2012).
Li, X. et al. The MOV10 helicase inhibits LINE-1 mobility. J. Biol. Chem. 288, 21148–21160 (2013).
Liu, Q. et al. MOV10 recruits DCP2 to decap human LINE-1 RNA by forming large cytoplasmic granules with phase separation properties. EMBO Rep. 24, e56512 (2023).
Li, X., Yu, H., Li, D. & Liu, N. LINE-1 transposable element renaissance in aging and age-related diseases. Ageing Res. Rev. 100, 102440 (2024).
Yue, Z. et al. Hyperglycaemia aggravates periodontal inflamm-aging by promoting SETDB1-mediated LINE-1 de-repression in macrophages. J. Clin. Periodontol. 50, 1685–1696 (2023).
Sookdeo, A., Hepp, C. M., McClure, M. A. & Boissinot, S. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4, 3 (2013).
Mangoni, D. et al. LINE-1 regulates cortical development by acting as long non-coding RNAs. Nat. Commun. 14, 4974 (2023).
Dewannieux, M. & Heidmann, T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J. Mol. Biol. 349, 241–247 (2005).
Raiz, J. et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683 (2012).
Zhang, B. et al. The senescence-associated secretory phenotype is potentiated by feedforward regulatory mechanisms involving Zscan4 and TAK1. Nat. Commun. 9, 1723 (2018).
Coppé, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Liu, H. et al. Rutin is a potent senomorphic agent to target senescent cells and can improve chemotherapeutic efficacy. Aging Cell 23, e13921 (2024).
Wiley, C. D. et al. Oxylipin biosynthesis reinforces cellular senescence and allows detection of senolysis. Cell Metab. 33, 1124–1136 (2021).
Czibik, G. et al. Dysregulated phenylalanine catabolism plays a key role in the trajectory of cardiac aging. Circulation 144, 559–574 (2021).
Bi, S. et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 11, 483–504 (2020).
Gulen, M. F. et al. cGAS–STING drives ageing-related inflammation and neurodegeneration. Nature 620, 374–380 (2023).
Haag, S. M. et al. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Lv, J. et al. Downregulation of MLF1 safeguards cardiomyocytes against senescence-associated chromatin opening. Nucleic Acids Res. 53, gkae1176 (2025).
Goodier, J. L., Cheung, L. E. & Kazazian, H. H. Jr. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 8, e1002941 (2012).
Choi, J., Hwang, S. Y. & Ahn, K. Interplay between RNASEH2 and MOV10 controls LINE-1 retrotransposition. Nucleic Acids Res. 46, 1912–1926 (2018).
Wei, J. et al. FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development. Science 376, 968–973 (2022).
Percharde, M. et al. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174, 391–405 (2018).
Solovyov, A. et al. Pan-cancer multi-omic model of LINE-1 activity reveals locus heterogeneity of retrotransposition efficiency. Nat. Commun. 16, 2049 (2025).
Takahashi, T. et al. LINE-1 activation in the cerebellum drives ataxia. Neuron 110, 3278–3287 (2022).
Liu, X. et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell 186, 287–304 (2023).
Zhang, Z. & Zhang, C. Regulation of cGAS–STING signalling and its diversity of cellular outcomes. Nat. Rev. Immunol. 25, 425–444 (2025).
Hu, D. et al. Cytosolic DNA sensor cGAS plays an essential pathogenetic role in pressure overload-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 318, H1525–h1537 (2020).
Cao, D. J. et al. Cytosolic DNA sensing promotes macrophage transformation and governs myocardial ischemic injury. Circulation 137, 2613–2634 (2018).
Luo, W. et al. Critical role of cytosolic DNA and its sensing adaptor STING in aortic degeneration, dissection, and rupture. Circulation 141, 42–66 (2020).
Wan, X. et al. cGAS–STING pathway performance in the vulnerable atherosclerotic plaque. Aging Dis. 13, 1606–1614 (2022).
Wang, J. et al. Prophylactic supplementation with Lactobacillus Reuteri or its metabolite GABA protects against acute ischemic cardiac injury. Adv. Sci. 11, e2307233 (2024).
Fan, Y. et al. ARID5A orchestrates cardiac aging and inflammation through MAVS mRNA stabilization. Nat. Cardiovasc. Res. 4, 602–623 (2025).
Xiao, Y. et al. Medium from human iPSC-derived primitive macrophages promotes adult cardiomyocyte proliferation and cardiac regeneration. Nat. Commun. 16, 3012 (2025).
Li, T. et al. Kansl1 haploinsufficiency impairs autophagosome-lysosome fusion and links autophagic dysfunction with Koolen-de Vries syndrome in mice. Nat. Commun. 13, 931 (2022).
Wang, Y. et al. Nynrin enhances cardiac function by inhibiting mitochondrial permeability transition pore opening upon myocardial ischemia/reperfusion injury. J. Mol. Cell. Cardiol. 209, 93–103 (2025).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wang, S. et al. Deciphering primate retinal aging at single-cell resolution. Protein Cell 12, 889–898 (2021).
Tu, C. et al. Targeting the chromatin remodeler BAZ2B mitigates hepatic senescence and MASH fibrosis. Nat. Aging 5, 1063–1078 (2025).
Cao, Q. et al. Targeting inflammation with chimeric antigen receptor macrophages using a signal switch. Nat. Biomed. Eng. 9, 1502–1516 (2025).
Mammoto, A., Muyleart, M. & Mammoto, T. LRP5 in age-related changes in vascular and alveolar morphogenesis in the lung. Aging 11, 89–103 (2019).
Wu, X. et al. Complement C3 deficiency ameliorates aging related changes in the kidney. Life Sci. 260, 118370 (2020).
Sovran, B. et al. Age-associated impairment of the mucus barrier function is associated with profound changes in microbiota and immunity. Sci Rep. 9, 1437 (2019).
Geng, L. et al. Systematic profiling reveals betaine as an exercise mimetic for geroprotection. Cell 188, 5403–5425 (2025).
Zhang, H. et al. Prophylactic supplementation with Bifidobacterium infantis or its metabolite inosine attenuates cardiac ischemia/reperfusion injury. Imeta 3, e220 (2024).
Wang, J. et al. CAR-macrophage therapy alleviates myocardial ischemia-reperfusion injury. Circ. Res. 135, 1161–1174 (2024).
Du, H. et al. CAR macrophages engineered in vivo for attenuating myocardial ischemia-reperfusion injury. Circ. Res. 137, 846–859 (2025).
Lei, J. et al. FOXO3-engineered human mesenchymal progenitor cells efficiently promote cardiac repair after myocardial infarction. Protein Cell 12, 145–151 (2021).
Du, H. et al. Flavonifractor Plautii or its metabolite desaminotyrosine as prophylactic agents for alleviating myocardial ischemia/reperfusion injury. Adv. Sci. 12, e2417827 (2025).
Zhang, Y. et al. Single-nucleus transcriptomics reveals a gatekeeper role for FOXP1 in primate cardiac aging. Protein Cell 14, 279–293 (2023).
Stack, E. C., Wang, C., Roman, K. A. & Hoyt, C. C. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70, 46–58 (2014).
Faget, L. & Hnasko, T. S. Tyramide signal amplification for immunofluorescent enhancement. Methods Mol. Biol. 1318, 161–172 (2015).
Seidlmayer, L. K. et al. Mitofusin 2 is essential for IP3-mediated SR/mitochondria metabolic feedback in ventricular myocytes. Front. Physiol. 10, 733 (2019).
Ackers-Johnson, M. et al. A simplified, langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ. Res. 119, 909–920 (2016).
Ye, Y. et al. SIRT2 counteracts primate cardiac aging via deacetylation of STAT3 that silences CDKN2B. Nat. Aging 3, 1269–1287 (2023).
Li, H. et al. Highly efficient generation of isogenic pluripotent stem cell models using prime editing. Elife 11, e79208 (2022).
Acknowledgements
We acknowledge Y. Hou for providing the Mov10loxP/loxP mouse strain. We thank Q. Xu from the Core Facilities Centre, Capital Medical University for technical support on echocardiography, and Z. Gao and J. Zang from the Institute of Zoology, Chinese Academy of Sciences for their administrative assistance. This work was supported by the National Natural Science Foundation of China (92368112), the Initiative Scientific Research Program of the Institute of Zoology, Chinese Academy of Sciences (2024IOZ0103 and 2023IOZ0202), the National Key Research and Development Program of China (2024YFA1802600), the CAS Project for Young Scientists in Basic Research (YSBR-076), the State Key Laboratory of Membrane Biology and the State Key Laboratory of Organ Regeneration and Reconstruction.
Author information
Authors and Affiliations
Contributions
C.Y. and H.D. performed and analyzed all experiments, except for those indicated below. S.L., Y.Z., J.S., X.Y., M.L., C.H., J.Z. and Y.X. performed histological studies. P.X. conducted RNA-sequencing analysis. Y.W. performed echocardiogram analysis. H.Y. and Y.L. conducted qPCR analysis. J.B. and Z.G. assisted with experiments. Y.H. contributed to developing mouse lines. J.Z. isolated primary cardiomyocytes. M.S. planned the study and designed the experiments. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Zhiyong Mao, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Assessment on transcript levels of transposable elements (TEs) during cardiac aging.
a, qPCR analysis showing LINE-1 ORF1 and ORF2 transcript levels in the hearts from 10-week, 30-week, 50-week, and 70-week wild-type mice. n = 5 mice per group. b, qPCR analysis showing SINE-B1 and SINE-B2 transcript levels in the hearts from each group. n = 5 mice per group. c, qPCR analysis showing MuLV, MMERVK10C, and MERVL transcript levels in the hearts from each group. n = 5 mice per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using one-way ANOVA and post hoc Dunnett′s test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
Extended Data Fig. 2 Cardiomyocyte-specific Mov10 knockout results in differential expression of transposable elements (TEs), diastolic dysfunction and cardiac senescence.
a, qPCR analysis showing SINE-B1 and SINE-B2 transcript levels in the hearts of littermate control (Ctrl) and Mov10-cKO (KO) mice at 3 months of age. n = 6 mice per group. b, qPCR analysis showing MuLV, MMERVK10C, and MERVL transcript levels in the hearts of Ctrl and KO mice at 3 months of age. n = 6 mice per group. c, Representative pulsed-wave Doppler (PWD, upper) and tissue Doppler imaging (TDI, lower) from Ctrl and KO mice at 3 months of age. Quantitative data on the ratio of early-diastolic transmitral flow velocity to early-diastolic mitral annular velocity (E/e′) are shown to the right. n = 7 mice per group. d, Masson’s trichrome staining of Ctrl and KO hearts at 3 months of age. Scale bar, 100 μm. Quantifications of cardiac fibrosis are shown to the right. n = 6 mice per group. e-f, qPCR analysis showing Cdkn1a and Cdkn2a (e), and Lmnb1 (f) mRNA levels in the hearts of Ctrl and KO mice at 3 months of age. n = 6 mice per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using unpaired two-tailed Student′s t-test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
Extended Data Fig. 3 Knockdown of LINE-1 alleviates senescence in Mov10-knockout H9C2 cells.
a, Immunoblotting analysis of LINE-1 ORF1p and ORF2p protein levels in wild-type (WT) and three independent lines of Mov10-knockout (KO-1, KO-2, and KO-3) H9C2 cells. GAPDH was used as a loading control. b, Quantification of LINE-1 ORF1p and ORF2p protein levels revealed by immunoblotting in WT and KO H9C2 cells transduced with Lenti-shCtrl or Lenti-shLINE-1. GAPDH was used as a loading control. n = 4 biological replicates per group. c, Immunoblotting analysis of p21 and p16 protein levels in WT and KO H9C2 cells transduced with Lenti-shCtrl or Lenti-shLINE-1. GAPDH was used as a loading control. n = 4 biological replicates per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using two-way ANOVA post hoc Tukey test. **p < 0.01, ***p < 0.001 compared to WT H9C2 cells transduced with Lenti-shCtrl; ns, not significant, #p < 0.05, ##p < 0.01, ###p < 0.001 compared to corresponding WT or KO lines transduced with Lenti-shCtrl.
Extended Data Fig. 4 Pharmacological inhibitors of LINE-1 reverse transcription or STING signaling attenuate senescence in Mov10-knockout H9C2 cells.
a-b, qPCR analysis showing Cdkn1a, Cdkn2a, and Lmnb1 mRNA levels in wild-type (WT) and three independent lines of Mov10-knockout (KO-1, KO-2, and KO-3) H9C2 cells treated with 3TC (a) or H-151 (b). n = 6 biological replicates per group. Groups were compared using two-way ANOVA post hoc Tukey test. ***p < 0.001 compared to WT H9C2 cells treated with PBS or DMSO as appropriate; ns, not significant, ###p < 0.001 compared to corresponding WT or KO lines treated with PBS or DMSO as appropriate.
Extended Data Fig. 5 MOV10 expression increases in mouse hearts with age.
a, Mov10 expression based on mouse cardiac aging RNA sequencing data from the GEO dataset GSE20074127. b, qPCR analysis of Mov10 mRNA expression in aged mouse heart tissue. n = 6 mice per group. c-d, Representative immunoblot (c) and quantitative analysis (d) of MOV10 protein levels in aged mouse hearts. GAPDH was used as a loading control. n = 4 mice per group. e, Scatter plots depicting the correlation of MOV10 with ORF1p and ORF2p with a fitted linear regression line (blue) and 95% confidence interval (gray shade). Statistical significance was determined by Pearson’s correlation, with the coefficient (R) and p value denoted for each pair. For panel a-b and d, Data are presented as mean ± SEM with individual data points indicated. Groups were compared using unpaired two-tailed Student′s t-test (a) or one-way ANOVA and post hoc Dunnett′s test (b, d). ns, not significant, ***p < 0.001.
Extended Data Fig. 6 Pharmacological inhibitors of LINE-1 reverse transcription or STING signaling exhibit no histological toxicity in major non-cardiac organs of aged mice.
a, Hematoxylin-eosin staining of liver, spleen, lung, kidney, and intestine from mice of 10 weeks old (10 wk), 48 weeks old (48 wk), 72 weeks old (72 wk), 72 weeks old and received 3TC treatment beginning at 48 weeks of age (72 wk + 3TC), and 72 weeks old and received H-151 treatment beginning at 48 weeks of age (72 wk + H-151). Scale bars, 200 μm. Histological injury was quantified as hepatic vacuolization area (%), splenic injury score, mean alveolar diameter, glomerular sclerosis index, and intestinal villus goblet-cell density. n = 6 mice per group. b, Measurement of serum levels of ALT (alanine aminotransferase), AST (aspartate aminotransferase), creatinine, and BUN (blood urea nitrogen). n = 5 mice per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using one-way ANOVA and post hoc Dunnett′s test except for the splenic injury score that was analyzed with the Kruskal-Wallis test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
Extended Data Fig. 7 Pharmacological inhibitors of LINE-1 reverse transcription or STING signaling attenuate cardiac aging phenotypes in aged mice.
a, Representative pulsed-wave Doppler (PWD, upper) and tissue Doppler imaging (TDI, lower) of mice of 10 weeks old (10 wk), 48 weeks old (48 wk), 72 weeks old (72 wk), 72 weeks old and received 3TC treatment beginning at 48 weeks of age (72 wk + 3TC), and 72 weeks old and received H-151 treatment beginning at 48 weeks of age (72 wk + H-151). Quantitative data on the ratio of early-diastolic transmitral flow velocity to early-diastolic mitral annular velocity (E/e′) are shown to the right. n = 5 mice for the first four groups and 3 for 72 wk + H-151 group. b, Relative LINE-1 DNA abundance in the hearts from each group. n = 6 mice per group. c, qPCR analysis showing LINE-1 ORF1 and ORF2 transcript levels in the hearts from each group. n = 5 mice per group. d, Heatmap showing transcript levels of SINE-B1, SINE-B2, MuLV, MMERVK10C, and MERVL in the hearts from each group. n = 5 mice per group. e, Immunohistochemical analysis of IL-1β (upper), TNF (middle), and F4/80 (lower) in the hearts from each group. Scale bar, 25 µm. Quantitative data on relative IL-1β and TNF signal intensities and the proportion of F4/80-positive cells are shown to the right. n = 6 mice per group. f, Immunohistochemical analysis of p21 (upper) and p16 (lower) in mouse hearts from each group. Brown color in the nuclei indicates the cells positive for p21 and p16. Scale bar, 25 µm. Quantitative data on the proportion of positive cells are shown to the right. n = 6 mice per group. Data are presented as mean ± SEM with individual data points indicated (a-c and e-f) or heatmap (d). Groups were compared using one-way ANOVA and post hoc Dunnett′s test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001 compared to 72 wk group.
Extended Data Fig. 8 Pharmacological inhibitor of LINE-1 reverse transcription improves cardiac function in Mov10-cKO mice.
a, Schematic for the experiment using pharmacological inhibition of LINE-1 reverse transcription (with 3TC) in alleviating premature cardiac aging and cardiac dysfunction in Mov10-cKO (KO) mice. KO and littermate control (Ctrl) mice received 3TC (administered in drinking water) or water alone from 7 weeks of age for 6 weeks before subsequent analyses. b, Representative images showing the M-mode (upper), pulsed-wave Doppler (PWD) (middle), and tissue Doppler imaging (TDI, lower) echocardiography of Ctrl mice that received water (Ctrl + Water), Ctrl mice that received 3TC (Ctrl + 3TC), Mov10-cKO mice that received water (KO + Water), Mov10-cKO mice that received 3TC (KO + 3TC). Quantitative data of left ventricular fractional shortening (LVFS), ejection fraction (LVEF), the ratio of E-wave to A-wave (E/A), the ratio of early-diastolic transmitral flow velocity to early-diastolic mitral annular velocity (E/e′) are shown to the right. n = 8 mice per group. c, Picrosirius red staining of mouse hearts from each group. Scale bar, 50 μm. Quantifications of cardiac fibrosis are shown to the right. n = 6 mice per group. d, Masson’s trichrome staining of mouse hearts from each group. Scale bar, 50 μm. Quantifications of cardiac fibrosis are shown to the right. n = 6 mice per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using two-way ANOVA post hoc Tukey test. ns, not significant, **p < 0.01, ***p < 0.001 compared to Ctrl + Water group; #p < 0.05, ###p < 0.001 compared to KO + Water group.
Extended Data Fig. 9 Pharmacological inhibitor of LINE-1 reverse transcription attenuates premature cardiac aging in Mov10-cKO mice.
a, Immunoblotting analysis of LINE-1 ORF1p and ORF2p protein levels in cardiomyocytes of Ctrl mice that received water (Ctrl + Water), Ctrl mice that received 3TC (Ctrl + 3TC), KO mice that received water (KO + Water), KO mice that received 3TC (KO + 3TC). GAPDH was used as a loading control. n = 4 mice per group. b, Immunoblotting analysis of p21 and p16 protein levels in the hearts from Ctrl mice that received water (Ctrl + Water), Ctrl mice that received 3TC (Ctrl + 3TC), KO mice that received water (KO + Water), KO mice that received 3TC (KO + 3TC). GAPDH was used as a loading control. n = 4 mice per group. c-d, Immunohistochemical analysis of p21 (c) and p16 (d) in mouse hearts from each group. Brown color in the nuclei indicates the cells positive for p21 and p16. Scale bar, 25 µm. n = 6 mice per group. e, SA-β-gal staining in mouse hearts from each group. Cyan color indicates cellular senescence. Scale bar, 50 μm. n = 6 mice per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using two-way ANOVA post hoc Tukey test. ns, not significant, ***p < 0.001 compared to Ctrl + Water group; #p < 0.05, ###p < 0.001 compared to KO + Water group.
Extended Data Fig. 10 Pharmacological inhibitor of LINE-1 reverse transcription mitigates cardiac inflammation in Mov10-cKO mice.
a–c, Immunohistochemical analysis of IL-1β (a), IL-6 (b), and TNF (c) in mouse hearts from each group. Scale bar, 25 µm. Quantitative data on relative signal intensities are shown to the right. n = 6 mice per group. Data are presented as mean ± SEM with individual data points indicated. Groups were compared using two-way ANOVA post hoc Tukey test. ns, not significant, ***p < 0.001 compared to Ctrl + Water group; #p < 0.05 compared to KO + Water group.
Supplementary information
Supplementary Table 1
Echocardiographic measurements.
Supplementary Table 2
Primers used for qPCR analysis.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, C., Du, H., Liu, S. et al. Targeting age-related LINE-1 activation alleviates cardiac aging. Nat Aging (2026). https://doi.org/10.1038/s43587-025-01056-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43587-025-01056-0